1ZZE
 
 | | X-ray Structure of NADPH-dependent Carbonyl Reductase from Sporobolomyces salmonicolor | | Descriptor: | Aldehyde reductase II, SULFATE ION | | Authors: | Kamitori, S, Iguchi, A, Ohtaki, A, Yamada, M, Kita, K. | | Deposit date: | 2005-06-14 | | Release date: | 2005-09-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray Structures of NADPH-dependent Carbonyl Reductase from Sporobolomyces salmonicolor Provide Insights into Stereoselective Reductions of Carbonyl Compounds
J.Mol.Biol., 352, 2005
|
|
2ZYK
 
 | | Crystal structure of cyclo/maltodextrin-binding protein complexed with gamma-cyclodextrin | | Descriptor: | Cyclooctakis-(1-4)-(alpha-D-glucopyranose), Solute-binding protein | | Authors: | Tonozuka, T, Sogawa, A, Yamada, M, Matsumoto, N, Yoshida, H, Kamitori, S, Ichikawa, K, Mizuno, M, Nishikawa, A, Sakano, Y. | | Deposit date: | 2009-01-26 | | Release date: | 2009-02-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for cyclodextrin recognition by Thermoactinomyces vulgaris cyclo/maltodextrin-binding protein
Febs J., 274, 2007
|
|
3WXC
 
 | | Crystal Structure of IMP-1 metallo-beta-lactamase complexed with a 3-aminophtalic acid inhibitor | | Descriptor: | 3-(4-hydroxypiperidin-1-yl)benzene-1,2-dicarboxylic acid, Beta-lactamase, ZINC ION | | Authors: | Saito, J, Watanabe, T, Yamada, M. | | Deposit date: | 2014-07-29 | | Release date: | 2014-10-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray crystallographic analysis of IMP-1 metallo-beta-lactamase complexed with a 3-aminophthalic acid derivative, structure-based drug design, and synthesis of 3,6-disubstituted phthalic acid derivative inhibitors
Bioorg.Med.Chem.Lett., 24, 2014
|
|
2ZBY
 
 | | Crystal structure of vitamin D hydroxylase cytochrome P450 105A1 (R84A mutant) | | Descriptor: | Cytochrome P450-SU1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugimoto, H, Shinkyo, R, Hayashi, K, Yoneda, S, Yamada, M, Kamakura, M, Ikushiro, S, Shiro, Y, Sakaki, T. | | Deposit date: | 2007-10-30 | | Release date: | 2008-04-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of CYP105A1 (P450SU-1) in Complex with 1alpha,25-Dihydroxyvitamin D3
Biochemistry, 47, 2008
|
|
2ZBX
 
 | | Crystal structure of vitamin D hydroxylase cytochrome P450 105A1 (wild type) with imidazole bound | | Descriptor: | Cytochrome P450-SU1, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugimoto, H, Shinkyo, R, Hayashi, K, Yoneda, S, Yamada, M, Kamakura, M, Ikushiro, S, Shiro, Y, Sakaki, T. | | Deposit date: | 2007-10-30 | | Release date: | 2008-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of CYP105A1 (P450SU-1) in Complex with 1alpha,25-Dihydroxyvitamin D3
Biochemistry, 47, 2008
|
|
2Z5M
 
 | | Complex of Transportin 1 with TAP NLS, crystal form 2 | | Descriptor: | Nuclear RNA export factor 1, Transportin-1 | | Authors: | Imasaki, T, Shimizu, T, Hashimoto, H, Hidaka, Y, Kose, S, Imamoto, N, Yamada, M, Sato, M. | | Deposit date: | 2007-07-14 | | Release date: | 2007-10-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for substrate recognition and dissociation by human transportin 1
Mol.Cell, 28, 2007
|
|
2ZBZ
 
 | | Crystal structure of vitamin D hydroxylase cytochrome P450 105A1 (R84A mutant) in complex with 1,25-dihydroxyvitamin D3 | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, Cytochrome P450-SU1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugimoto, H, Shinkyo, R, Hayashi, K, Yoneda, S, Yamada, M, Kamakura, M, Ikushiro, S, Shiro, Y, Sakaki, T. | | Deposit date: | 2007-10-30 | | Release date: | 2008-04-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of CYP105A1 (P450SU-1) in Complex with 1alpha,25-Dihydroxyvitamin D3
Biochemistry, 47, 2008
|
|
2Z5N
 
 | | Complex of Transportin 1 with hnRNP D NLS | | Descriptor: | Heterogeneous nuclear ribonucleoprotein D0, Transportin-1 | | Authors: | Imasaki, T, Shimizu, T, Hashimoto, H, Hidaka, Y, Kose, S, Imamoto, N, Yamada, M, Sato, M. | | Deposit date: | 2007-07-14 | | Release date: | 2007-10-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for substrate recognition and dissociation by human transportin 1
Mol.Cell, 28, 2007
|
|
2Z5J
 
 | | Free Transportin 1 | | Descriptor: | Transportin-1 | | Authors: | Imasaki, T, Shimizu, T, Hashimoto, H, Hidaka, Y, Yamada, M, Sato, M. | | Deposit date: | 2007-07-13 | | Release date: | 2007-10-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis for substrate recognition and dissociation by human transportin 1
Mol.Cell, 28, 2007
|
|
2Z5O
 
 | | Complex of Transportin 1 with JKTBP NLS | | Descriptor: | Heterogeneous nuclear ribonucleoprotein D-like, Transportin-1 | | Authors: | Imasaki, T, Shimizu, T, Hashimoto, H, Hidaka, Y, Kose, S, Imamoto, N, Yamada, M, Sato, M. | | Deposit date: | 2007-07-14 | | Release date: | 2007-10-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for substrate recognition and dissociation by human transportin 1
Mol.Cell, 28, 2007
|
|
3APM
 
 | | Crystal structure of the human SNP PAD4 protein | | Descriptor: | Protein-arginine deiminase type-4 | | Authors: | Horikoshi, N, Tachiwana, H, Saito, K, Osakabe, A, Sato, M, Yamada, M, Akashi, S, Nishimura, Y, Kagawa, W, Kurumizaka, H. | | Deposit date: | 2010-10-19 | | Release date: | 2011-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and biochemical analyses of the human PAD4 variant encoded by a functional haplotype gene
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3APN
 
 | | Crystal structure of the human wild-type PAD4 protein | | Descriptor: | Protein-arginine deiminase type-4 | | Authors: | Horikoshi, N, Tachiwana, H, Saito, K, Osakabe, A, Sato, M, Yamada, M, Akashi, S, Nishimura, Y, Kagawa, W, Kurumizaka, H. | | Deposit date: | 2010-10-19 | | Release date: | 2011-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and biochemical analyses of the human PAD4 variant encoded by a functional haplotype gene
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2Z5K
 
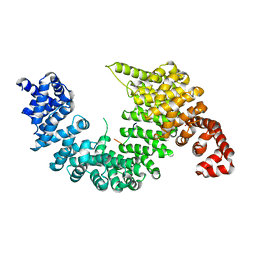 | | Complex of Transportin 1 with TAP NLS | | Descriptor: | Nuclear RNA export factor 1, PHOSPHATE ION, Transportin-1 | | Authors: | Imasaki, T, Shimizu, T, Hashimoto, H, Hidaka, Y, Yamada, M, Kose, S, Imamoto, N, Sato, M. | | Deposit date: | 2007-07-14 | | Release date: | 2007-10-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for substrate recognition and dissociation by human transportin 1
Mol.Cell, 28, 2007
|
|
2Z6J
 
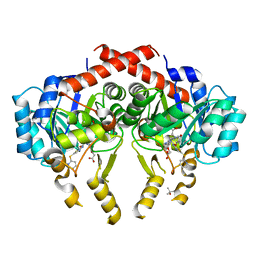 | | Crystal Structure of S. pneumoniae Enoyl-Acyl Carrier Protein Reductase (FabK) in Complex with an Inhibitor | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(4-(2-((3-(5-(PYRIDIN-2-YLTHIO)THIAZOL-2-YL)UREIDO)METHYL)-1H-IMIDAZOL-4-YL)PHENOXY)ACETIC ACID, CALCIUM ION, ... | | Authors: | Saito, J, Yamada, M, Watanabe, T, Takeuchi, Y. | | Deposit date: | 2007-08-01 | | Release date: | 2008-04-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of enoyl-acyl carrier protein reductase (FabK) from Streptococcus pneumoniae reveals the binding mode of an inhibitor.
Protein Sci., 17, 2008
|
|
2Z6I
 
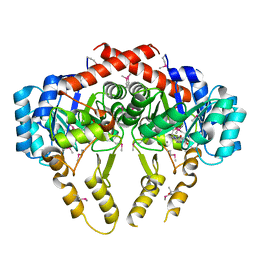 | | Crystal Structure of S. pneumoniae Enoyl-Acyl Carrier Protein Reductase (FabK) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Saito, J, Yamada, M, Watanabe, T, Takeuchi, Y. | | Deposit date: | 2007-08-01 | | Release date: | 2008-04-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of enoyl-acyl carrier protein reductase (FabK) from Streptococcus pneumoniae reveals the binding mode of an inhibitor.
Protein Sci., 17, 2008
|
|
2ZYN
 
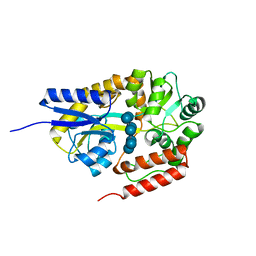 | | Crystal structure of cyclo/maltodextrin-binding protein complexed with beta-cyclodextrin | | Descriptor: | Cycloheptakis-(1-4)-(alpha-D-glucopyranose), Solute-binding protein | | Authors: | Matsumoto, M, Yamada, M, Kurakata, Y, Yoshida, H, Kamitori, S, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2009-01-27 | | Release date: | 2009-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of open and closed forms of cyclo/maltodextrin-binding protein
Febs J., 276, 2009
|
|
2ZYO
 
 | | Crystal structure of cyclo/maltodextrin-binding protein complexed with maltotetraose | | Descriptor: | alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, solute-binding protein | | Authors: | Matsumoto, M, Yamada, M, Kurakata, Y, Yoshida, H, Kamitori, S, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2009-01-27 | | Release date: | 2009-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of open and closed forms of cyclo/maltodextrin-binding protein
Febs J., 276, 2009
|
|
2ZYM
 
 | | Crystal structure of cyclo/maltodextrin-binding protein complexed with alpha-cyclodextrin | | Descriptor: | Cyclohexakis-(1-4)-(alpha-D-glucopyranose), Solute-binding protein | | Authors: | Matsumoto, M, Yamada, M, Kurakata, Y, Yoshida, H, Kamitori, S, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2009-01-27 | | Release date: | 2009-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of open and closed forms of cyclo/maltodextrin-binding protein
Febs J., 276, 2009
|
|
