1YMW
 
 | | The study of reductive unfolding pathways of RNase A (Y92G mutant) | | 分子名称: | Ribonuclease pancreatic | | 著者 | Xu, G, Narayan, M, Kurinov, I, Ripoll, D.R, Welker, E, Khalili, M, Ealick, S.E, Scheraga, H.A. | | 登録日 | 2005-01-21 | | 公開日 | 2006-01-31 | | 最終更新日 | 2021-10-20 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | A localized specific interaction alters the unfolding pathways of structural homologues.
J.Am.Chem.Soc., 128, 2006
|
|
1YMR
 
 | | The study of reductive unfolding pathways of RNase A (Y92A mutant) | | 分子名称: | Ribonuclease pancreatic | | 著者 | Xu, G, Narayan, M, Kurinov, I, Ripoll, D.R, Welker, E, Khalili, M, Ealick, S.E, Scheraga, H.A. | | 登録日 | 2005-01-21 | | 公開日 | 2006-01-31 | | 最終更新日 | 2021-10-20 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | A localized specific interaction alters the unfolding pathways of structural homologues.
J.Am.Chem.Soc., 128, 2006
|
|
1YMN
 
 | | The study of reductive unfolding pathways of RNase A (Y92L mutant) | | 分子名称: | Ribonuclease pancreatic | | 著者 | Xu, G, Narayan, M, Kurinov, I, Ripoll, D.R, Welker, E, Khalili, M, Ealick, S.E, Scheraga, H.A. | | 登録日 | 2005-01-21 | | 公開日 | 2006-01-31 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | A localized specific interaction alters the unfolding pathways of structural homologues.
J.Am.Chem.Soc., 128, 2006
|
|
3RZF
 
 | | Crystal Structure of Inhibitor of kappaB kinase beta (I4122) | | 分子名称: | (4-{[4-(4-chlorophenyl)pyrimidin-2-yl]amino}phenyl)[4-(2-hydroxyethyl)piperazin-1-yl]methanone, MGC80376 protein | | 著者 | Xu, G, Lo, Y.C, Li, Q, Napolitano, G, Wu, X, Jiang, X, Dreano, M, Karin, M, Wu, H. | | 登録日 | 2011-05-11 | | 公開日 | 2011-05-25 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (4 Å) | | 主引用文献 | Crystal structure of inhibitor of KappaB kinase Beta.
Nature, 472, 2011
|
|
7W9N
 
 | | THE STRUCTURE OF OBA33-OTA COMPLEX | | 分子名称: | (2~{S})-2-[[(3~{R})-5-chloranyl-3-methyl-8-oxidanyl-1-oxidanylidene-3,4-dihydroisochromen-7-yl]carbonylamino]-3-phenyl-propanoic acid, OTA DNA APTAMER (33-MER) | | 著者 | Xu, G.H, Li, C.G. | | 登録日 | 2021-12-10 | | 公開日 | 2022-01-19 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Insights into the Mechanism of High-Affinity Binding of Ochratoxin A by a DNA Aptamer.
J.Am.Chem.Soc., 144, 2022
|
|
5WLS
 
 | | Crystal Structure of a Pollen Receptor Kinase 3 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Pollen receptor-like kinase 3 | | 著者 | Xu, G, Chakraborty, S, Pan, H. | | 登録日 | 2017-07-27 | | 公開日 | 2018-06-06 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.496 Å) | | 主引用文献 | The Extracellular Domain of Pollen Receptor Kinase 3 is structurally similar to the SERK family of co-receptors.
Sci Rep, 8, 2018
|
|
7W3U
 
 | | USP34 catalytic domain in complex with UbPA | | 分子名称: | Polyubiquitin-B, Ubiquitin carboxyl-terminal hydrolase 34, ZINC ION, ... | | 著者 | Xu, G.L, Ming, Z.H. | | 登録日 | 2021-11-26 | | 公開日 | 2022-06-01 | | 最終更新日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (3.13 Å) | | 主引用文献 | Structural Insights into the Catalytic Mechanism and Ubiquitin Recognition of USP34.
J.Mol.Biol., 434, 2022
|
|
7W3R
 
 | | USP34 catalytic domain | | 分子名称: | Ubiquitin carboxyl-terminal hydrolase 34, ZINC ION | | 著者 | Xu, G.L, Ming, Z.H. | | 登録日 | 2021-11-26 | | 公開日 | 2022-06-01 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Structural Insights into the Catalytic Mechanism and Ubiquitin Recognition of USP34.
J.Mol.Biol., 434, 2022
|
|
7WQO
 
 | |
7WQP
 
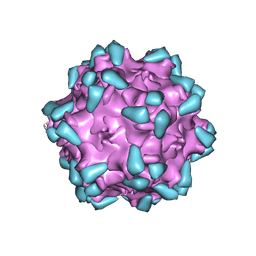 | | Adeno-associated virus serotype PHP.eB in complex with AAVR | | 分子名称: | Capsid protein VP1, Dyslexia-associated protein KIAA0319-like protein | | 著者 | Xu, G, Lou, Z. | | 登録日 | 2022-01-25 | | 公開日 | 2022-06-15 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (3.76 Å) | | 主引用文献 | Structural basis for the neurotropic AAV9 and the engineered AAVPHP.eB recognition with cellular receptors.
Mol Ther Methods Clin Dev, 26, 2022
|
|
7WJW
 
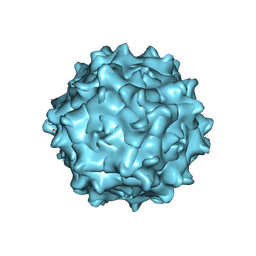 | | Structure of Adeno-associated virus serotype 9 | | 分子名称: | Capsid protein VP1 | | 著者 | Xu, G, Lou, Z. | | 登録日 | 2022-01-08 | | 公開日 | 2022-06-15 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (3.87 Å) | | 主引用文献 | Structural basis for the neurotropic AAV9 and the engineered AAVPHP.eB recognition with cellular receptors.
Mol Ther Methods Clin Dev, 26, 2022
|
|
7WJX
 
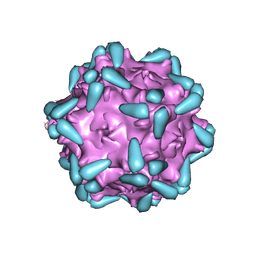 | | Adeno-associated virus serotype 9 in complex with AAVR | | 分子名称: | Capsid protein VP1, Dyslexia-associated protein KIAA0319-like protein | | 著者 | Xu, G, Lou, Z. | | 登録日 | 2022-01-08 | | 公開日 | 2022-06-15 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (3.23 Å) | | 主引用文献 | Structural basis for the neurotropic AAV9 and the engineered AAVPHP.eB recognition with cellular receptors.
Mol Ther Methods Clin Dev, 26, 2022
|
|
7YMU
 
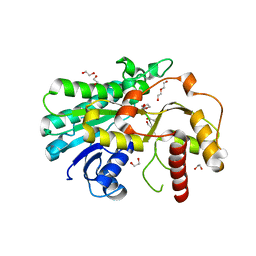 | | Structure of Alcohol dehydrogenase from [Candida] glabrata | | 分子名称: | 1,2-ETHANEDIOL, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Sun, Z.W, Liu, Y.F, Xu, G.C, Ni, Y. | | 登録日 | 2022-07-29 | | 公開日 | 2023-08-02 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Rationla design of CgADH from candida glarata for asymmetric reduction of azacycolne.
To Be Published
|
|
8HXK
 
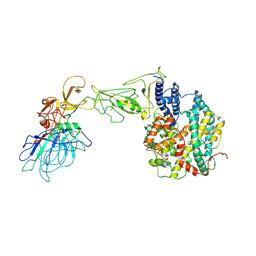 | | BANAL-20-236 S1 in complex with R. Affinis ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | 著者 | Wang, X, Xu, G. | | 登録日 | 2023-01-04 | | 公開日 | 2024-01-10 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | The selective effect of fecal-oral transmission on the S proteins of bat SARS-CoV-2 related coronaviruses in favor of stability over infectivity
To Be Published
|
|
7YMB
 
 | |
8I99
 
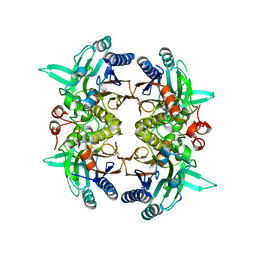 | | N-carbamoyl-D-amino-acid hydrolase mutant - M4Th3 | | 分子名称: | N-carbamoyl-D-amino-acid hydrolase | | 著者 | Hu, J.M, Ni, Y, Xu, G.C. | | 登録日 | 2023-02-06 | | 公開日 | 2023-03-22 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (3.17 Å) | | 主引用文献 | Engineering the Thermostability of a d-Carbamoylase Based on Ancestral Sequence Reconstruction for the Efficient Synthesis of d-Tryptophan.
J.Agric.Food Chem., 71, 2023
|
|
8HXJ
 
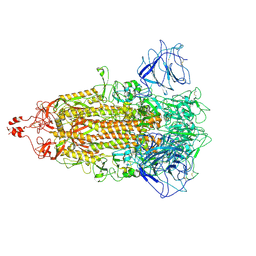 | | BANAL-20-52 Spike trimer | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Wang, X, Xu, G. | | 登録日 | 2023-01-04 | | 公開日 | 2024-01-10 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | The selective effect of fecal-oral transmission on the S proteins of bat SARS-CoV-2 related coronaviruses in favor of stability over infectivity
To Be Published
|
|
6LED
 
 | |
6LE2
 
 | | Structure of D-carbamoylase mutant from Nitratireductor indicus | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, N-carbamoyl-D-amino-acid hydrolase | | 著者 | Ni, Y, Liu, Y.F, Xu, G.C, Dai, W. | | 登録日 | 2019-11-23 | | 公開日 | 2020-10-28 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | Structure-Guided Engineering of D-Carbamoylase Reveals a Key Loop at Substrate Entrance Tunnel
Acs Catalysis, 10, 2020
|
|
2VK7
 
 | | THE STRUCTURE OF CLOSTRIDIUM PERFRINGENS NANI SIALIDASE AND ITS CATALYTIC INTERMEDIATES | | 分子名称: | 5-acetamido-3,5-dideoxy-3-fluoro-D-erythro-alpha-L-manno-non-2-ulopyranosonic acid, CALCIUM ION, EXO-ALPHA-SIALIDASE | | 著者 | Newstead, S.L, Potter, J.A, Wilson, J.C, Xu, G, Chien, C.H, Watts, A.G, Withers, S.G, Taylor, G.L. | | 登録日 | 2007-12-17 | | 公開日 | 2008-01-22 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | The Structure of Clostridium Perfringens Nani Sialidase and its Catalytic Intermediates.
J.Biol.Chem., 283, 2008
|
|
2VK6
 
 | | THE STRUCTURE OF CLOSTRIDIUM PERFRINGENS NANI SIALIDASE AND ITS CATALYTIC INTERMEDIATES | | 分子名称: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, CALCIUM ION, EXO-ALPHA-SIALIDASE, ... | | 著者 | Newstead, S.L, Potter, J.A, Wilson, J.C, Xu, G, Chien, C.H, Watts, A.G, Withers, S.G, Taylor, G.L. | | 登録日 | 2007-12-17 | | 公開日 | 2008-01-22 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | The Structure of Clostridium Perfringens Nani Sialidase and its Catalytic Intermediates.
J.Biol.Chem., 283, 2008
|
|
2VK5
 
 | | THE STRUCTURE OF CLOSTRIDIUM PERFRINGENS NANI SIALIDASE AND ITS CATALYTIC INTERMEDIATES | | 分子名称: | CALCIUM ION, EXO-ALPHA-SIALIDASE, GLYCEROL | | 著者 | Newstead, S.L, Potter, J.A, Wilson, J.C, Xu, G, Chien, C.H, Watts, A.G, Withers, S.G, Taylor, G.L. | | 登録日 | 2007-12-17 | | 公開日 | 2008-01-22 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (0.97 Å) | | 主引用文献 | The Structure of Clostridium Perfringens Nani Sialidase and its Catalytic Intermediates.
J.Biol.Chem., 283, 2008
|
|
2XCY
 
 | | Crystal structure of Aspergillus fumigatus sialidase | | 分子名称: | CHLORIDE ION, EXTRACELLULAR SIALIDASE/NEURAMINIDASE, PUTATIVE, ... | | 著者 | Telford, J.C, Yeung, J, Xu, G, Bennet, A, Moore, M.M, Taylor, G.L. | | 登録日 | 2010-04-27 | | 公開日 | 2010-05-12 | | 最終更新日 | 2017-06-28 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | The Aspergillus Fumigatus Sialidase is a Kdnase: Structural and Mechanistic Insights.
J.Biol.Chem., 286, 2011
|
|
2YA5
 
 | | Crystal structure of Streptococcus pneumoniae NanA (TIGR4) in complex with sialic acid | | 分子名称: | CHLORIDE ION, FORMIC ACID, N-acetyl-alpha-neuraminic acid, ... | | 著者 | Gut, H, Xu, G, Taylor, G.L, Walsh, M.A. | | 登録日 | 2011-02-18 | | 公開日 | 2011-04-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural Basis for Streptococcus Pneumoniae Nana Inhibition by Influenza Antivirals Zanamivir and Oseltamivir Carboxylate.
J.Mol.Biol., 409, 2011
|
|
2YA7
 
 | | Crystal structure of Streptococcus pneumoniae NanA (TIGR4) in complex with Zanamivir | | 分子名称: | CHLORIDE ION, NEURAMINIDASE A, ZANAMIVIR | | 著者 | Gut, H, Xu, G, Taylor, G.L, Walsh, M.A. | | 登録日 | 2011-02-18 | | 公開日 | 2011-04-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Structural Basis for Streptococcus Pneumoniae Nana Inhibition by Influenza Antivirals Zanamivir and Oseltamivir Carboxylate.
J.Mol.Biol., 409, 2011
|
|
