8WY0
 
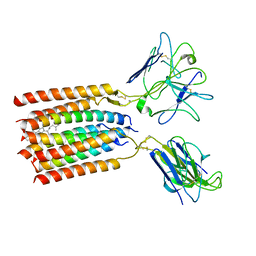 | | T cell receptor delta 2 gamma 9 with F283A, F290A, and F291A | | Descriptor: | CHOLESTEROL, Signal peptide,flag tag,T cell receptor delta variable 2,T cell receptor delta constant, Signal peptide,flag tag,T cell receptor gamma variable 9,T cell receptor gamma constant 1, ... | | Authors: | Xin, W, Huang, B, Chi, X, Liu, Y, Xu, M, Zhang, Y, Li, X, Su, Q, Zhou, Q. | | Deposit date: | 2023-10-30 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of human gamma delta T cell receptor-CD3 complex.
Nature, 630, 2024
|
|
8WYI
 
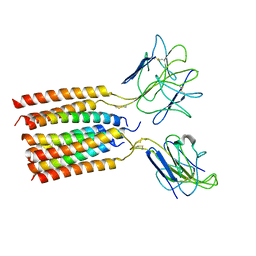 | | T cell receptor delta 2 gamma 9 with TCRD TM domain chimera of TRAC | | Descriptor: | Signal peptide,flag tag,T cell receptor delta variable 2,T cell receptor delta constant,T cell receptor alpha chain constant,T cell receptor delta variable 2,T cell receptor delta constant,T cell receptor alpha chain constant, Signal peptide,flag tag,T cell receptor gamma variable 9,T cell receptor gamma constant 1,T cell receptor gamma variable 9,T cell receptor gamma constant 1, T-cell surface glycoprotein CD3 delta chain, ... | | Authors: | Xin, W, Huang, B, Chi, X, Liu, Y, Xu, M, Zhang, Y, Li, X, Su, Q, Zhou, Q. | | Deposit date: | 2023-10-31 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of human gamma delta T cell receptor-CD3 complex.
Nature, 630, 2024
|
|
8WXE
 
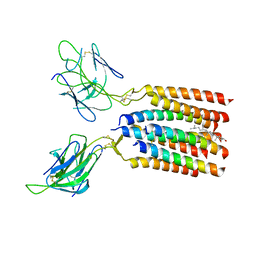 | | Vgamma5Vdelta1 EH TCR-CD3 complex | | Descriptor: | CHOLESTEROL, Signal peptide,flag tag,T cell receptor delta variable 1,T cell receptor delta constant, Signal peptide,flag tag,T cell receptor gamma variable 5,T cell receptor gamma constant 1, ... | | Authors: | Xin, W, Huang, B, Chi, X, Xu, M, Zhang, Y, Li, X, Su, Q, Zhou, Q. | | Deposit date: | 2023-10-28 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of human gamma delta T cell receptor-CD3 complex.
Nature, 630, 2024
|
|
8YC0
 
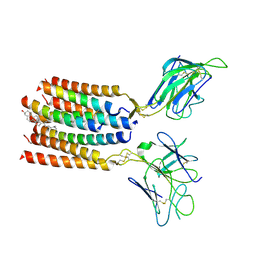 | | T cell receptor V delta2 V gamma9 in GDN | | Descriptor: | CHOLESTEROL, T cell receptor delta variable 2,T cell receptor delta constant, T cell receptor gamma variable 9,T cell receptor gamma constant 1, ... | | Authors: | Xin, W, Huang, B, Chi, X, Xu, M, Zhang, Y, Li, X, Su, Q, Zhou, Q. | | Deposit date: | 2024-02-17 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.12 Å) | | Cite: | Structures of human gamma delta T cell receptor-CD3 complex.
Nature, 630, 2024
|
|
8JCB
 
 | | Vgamma5 Vdelta1 T cell receptor complex | | Descriptor: | T cell receptor delta variable 1,T cell receptor delta constant, T cell receptor gamma variable 5,T cell receptor gamma constant 1, T-cell surface glycoprotein CD3 delta chain, ... | | Authors: | Xin, W, Huang, B, Chi, X, Xu, M, Zhang, Y, Li, X, Su, Q, Zhou, Q. | | Deposit date: | 2023-05-10 | | Release date: | 2024-05-08 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Structures of human gamma delta T cell receptor-CD3 complex.
Nature, 630, 2024
|
|
8JBV
 
 | | Extracellular domain of gamma delta TCR | | Descriptor: | T cell receptor delta variable 1,T cell receptor delta constant, T cell receptor gamma variable 5,T cell receptor gamma constant 1 | | Authors: | Xin, W, Chi, X, Huang, B, Su, Q, Zhou, Q. | | Deposit date: | 2023-05-09 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structures of human gamma delta T cell receptor-CD3 complex.
Nature, 630, 2024
|
|
8JC0
 
 | | V gamma9 V delta2 TCR and CD3 complex in LMNG | | Descriptor: | CHOLESTEROL, T cell receptor delta variable 2,T cell receptor delta constant, T cell receptor gamma variable 9,T cell receptor gamma constant 1, ... | | Authors: | Xin, W, Huang, B, Chi, X, Xu, M, Zhang, Y, Li, X, Su, Q, Zhou, Q. | | Deposit date: | 2023-05-10 | | Release date: | 2024-05-08 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of human gamma delta T cell receptor-CD3 complex.
Nature, 630, 2024
|
|
2QKW
 
 | | Structural basis for activation of plant immunity by bacterial effector protein AvrPto | | Descriptor: | Avirulence protein, Protein kinase | | Authors: | Xing, W.M, Zou, Y, Liu, Q, Hao, Q, Zhou, J.M, Chai, J.J. | | Deposit date: | 2007-07-11 | | Release date: | 2007-08-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structural basis for activation of plant immunity by bacterial effector protein AvrPto
Nature, 449, 2007
|
|
7XP9
 
 | | Phytophthora infesfans RxLR effector AVRvnt1 | | Descriptor: | RxLR effector protein Avr-vnt11 | | Authors: | Xing, W, Hu, Q, Zhou, J, Yao, D. | | Deposit date: | 2022-05-04 | | Release date: | 2023-06-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Chloroplast Protein GLYK Hijacked by Phytophthora Infestans Effector AVRvnt1 in Cytoplasm to Activate NLR
To Be Published
|
|
8ZJB
 
 | |
7C96
 
 | | Avr1d:GmPUB13 U-box | | Descriptor: | RING-type E3 ubiquitin transferase, RxLR effector protein Avh6 | | Authors: | Xing, W, Hu, Q, Zhou, J, Yao, D. | | Deposit date: | 2020-06-05 | | Release date: | 2021-03-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Phytophthora sojae effector Avr1d functions as an E2 competitor and inhibits ubiquitination activity of GmPUB13 to facilitate infection.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4I6E
 
 | | A vertebrate cryptochrome | | Descriptor: | Cryptochrome-2 | | Authors: | Xing, W, Busino, L, Hinds, T.R, Marionni, S.T, Saifee, N.H, Bush, M.F, Pagano, M, Zheng, N. | | Deposit date: | 2012-11-29 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | SCFFBXL3 ubiquitin ligase targets cryptochromes at their cofactor pocket.
Nature, 496, 2013
|
|
4I6G
 
 | | a vertebrate cryptochrome with FAD | | Descriptor: | Cryptochrome-2, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Xing, W, Busino, L, Hinds, T.R, Marionni, S.T, Saifee, N.H, Bush, M.F, Pagano, M, Zheng, N. | | Deposit date: | 2012-11-29 | | Release date: | 2013-03-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | SCFFBXL3 ubiquitin ligase targets cryptochromes at their cofactor pocket.
Nature, 496, 2013
|
|
4I6J
 
 | | A ubiquitin ligase-substrate complex | | Descriptor: | Cryptochrome-2, F-box/LRR-repeat protein 3, S-phase kinase-associated protein 1 | | Authors: | Xing, W, Busino, L, Hinds, T.R, Marionni, S.T, Saifee, N.H, Bush, M.F, Pagano, M, Zheng, N. | | Deposit date: | 2012-11-29 | | Release date: | 2013-03-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | SCFFBXL3 ubiquitin ligase targets cryptochromes at their cofactor pocket.
Nature, 496, 2013
|
|
7X8U
 
 | |
1R33
 
 | | Golgi alpha-mannosidase II complex with 5-thio-D-mannopyranosylamine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-thio-alpha-D-mannopyranosylamine, ... | | Authors: | Kuntz, D.A, Xin, W, Kavelekar, L.M, Rose, D.R, Pinto, B.M. | | Deposit date: | 2003-09-30 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 5-Thio-d-glycopyranosylamines and their amidinium salts as potential transition-state mimics of glycosyl hydrolases: synthesis, enzyme inhibitory activities, X-ray crystallography, and molecular modeling
TETRAHEDRON ASYMMETRY, 16, 2005
|
|
1R34
 
 | | Golgi alpha-mannosidase II complex with 5-thio-D-mannopyranosylamidinium salt | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, N-[(1Z)-2-phenylethanimidoyl]-5-thio-alpha-D-mannopyranosylamine, ... | | Authors: | Kuntz, D.A, Xin, W, Kavelekar, L.M, Rose, D.R, Pinto, B.M. | | Deposit date: | 2003-09-30 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 5-Thio-d-glycopyranosylamines and their amidinium salts as potential transition-state mimics of glycosyl hydrolases: synthesis, enzyme inhibitory activities, X-ray crystallography, and molecular modeling
TETRAHEDRON ASYMMETRY, 16, 2005
|
|
7XPC
 
 | |
4MLP
 
 | |
7E40
 
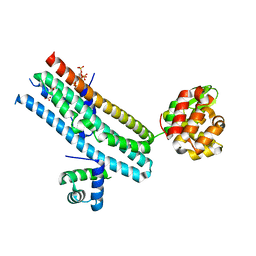 | | Mechanism of Phosphate Sensing and Signaling Revealed by Rice SPX1-PHR2 Complex Structure | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Protein PHOSPHATE STARVATION RESPONSE 2, SPX domain-containing protein 1,Endolysin | | Authors: | Zhou, J, Hu, Q, Yao, D, Xing, W. | | Deposit date: | 2021-02-09 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mechanism of phosphate sensing and signaling revealed by rice SPX1-PHR2 complex structure.
Nat Commun, 12, 2021
|
|
7VS2
 
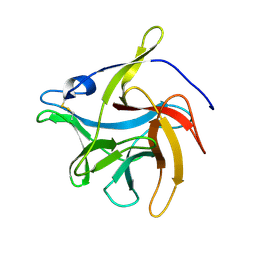 | | secreted fungal effector protein MoErs1 | | Descriptor: | MoErs1 | | Authors: | Wang, F.F, Xing, W.M. | | Deposit date: | 2021-10-25 | | Release date: | 2023-08-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Targeting Magnaporthe oryzae effector MoErs1 and host papain-like protease OsRD21 interaction to combat rice blast.
Nat.Plants, 2024
|
|
6LIX
 
 | | CRL Protein of Arabidopsis | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Chromophore lyase CRL, chloroplastic | | Authors: | Wang, F.F, Guan, K.L, Sun, P.K, Xing, W.M. | | Deposit date: | 2019-12-13 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.385 Å) | | Cite: | The Arabidopsis CRUMPLED LEAF protein, a homolog of the cyanobacterial bilin lyase, retains the bilin-binding pocket for a yet unknown function.
Plant J., 104, 2020
|
|
6LIY
 
 | | SeMet CRL Protein of Arabidopsis | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Chromophore lyase CRL, chloroplastic | | Authors: | Wang, F.F, Guan, K.L, Sun, P.K, Xing, W.M. | | Deposit date: | 2019-12-13 | | Release date: | 2020-09-16 | | Last modified: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (1.761 Å) | | Cite: | The Arabidopsis CRUMPLED LEAF protein, a homolog of the cyanobacterial bilin lyase, retains the bilin-binding pocket for a yet unknown function.
Plant J., 104, 2020
|
|
7XA9
 
 | | Structure of Arabidopsis thaliana CLCa | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chloride channel protein CLC-a, MAGNESIUM ION, ... | | Authors: | Ji, S, Jin, H, Kaiming, Z, Mingxing, W, Shanshan, L, Long, C. | | Deposit date: | 2022-03-17 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Cryo-EM structure of the plant nitrate transporter AtCLCa reveals characteristics of the anion-binding site and the ATP-binding pocket.
J.Biol.Chem., 299, 2023
|
|
