5H5D
 
 | |
5X3G
 
 | |
5X3H
 
 | |
8HVJ
 
 | |
5YAC
 
 | | Crystal structure of WT Trm5b from Pyrococcus abyssi | | Descriptor: | tRNA (guanine(37)-N1)-methyltransferase Trm5b | | Authors: | Xie, W, Wu, J. | | Deposit date: | 2017-08-31 | | Release date: | 2017-09-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The crystal structure of the Pyrococcus abyssi mono-functional methyltransferase PaTrm5b
Biochem. Biophys. Res. Commun., 493, 2017
|
|
5Y9O
 
 | | The apo structure of Legionella pneumophila WipA | | Descriptor: | IODIDE ION, WipA | | Authors: | Xie, W, Jia, Q. | | Deposit date: | 2017-08-26 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.554 Å) | | Cite: | Legionella pneumophila effector WipA, a bacterial PPP protein phosphatase with PTP activity
Acta Biochim. Biophys. Sin. (Shanghai), 50, 2018
|
|
6A6X
 
 | | The crystal structure of the Mtb MazE-MazF-mt9 complex | | Descriptor: | Antitoxin MazE7, Probable endoribonuclease MazF7, SULFATE ION | | Authors: | Xie, W, Chen, R, Tu, J. | | Deposit date: | 2018-06-29 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Biochemical Characterization of the Cognate and Heterologous Interactions of the MazEF-mt9 TA System.
Acs Infect Dis., 5, 2019
|
|
5XE0
 
 | | Crystal structure of EV-D68-3Dpol in complex with GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Genome polyprotein | | Authors: | Xie, W, Wang, C, Wang, Z, Li, Q, Wang, C. | | Deposit date: | 2017-03-30 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure and Thermostability Characterization of Enterovirus D68 3Dpol
J. Virol., 91, 2017
|
|
5X62
 
 | |
6IOD
 
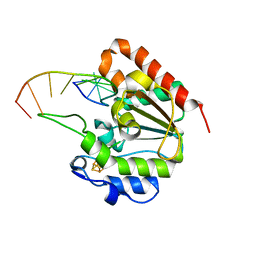 | | The structure of UdgX in complex with single-stranded DNA | | Descriptor: | DNA, IRON/SULFUR CLUSTER, Phage SPO1 DNA polymerase-related protein | | Authors: | Xie, W, Tu, J. | | Deposit date: | 2018-10-29 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Suicide inactivation of the uracil DNA glycosylase UdgX by covalent complex formation.
Nat.Chem.Biol., 15, 2019
|
|
6JFP
 
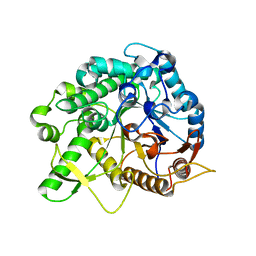 | | Crystal structure of the beta-glucosidase Bgl15 | | Descriptor: | beta-D-glucopyranose, beta-glucosidase 15 | | Authors: | Xie, W, Chen, R. | | Deposit date: | 2019-02-11 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Engineering of beta-Glucosidase Bgl15 with Simultaneously Enhanced Glucose Tolerance and Thermostability To Improve Its Performance in High-Solid Cellulose Hydrolysis.
J.Agric.Food Chem., 68, 2020
|
|
7VWN
 
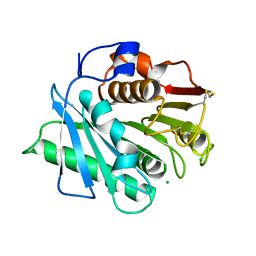 | | The structure of an engineered PET hydrolase | | Descriptor: | CHLORIDE ION, Poly(ethylene terephthalate) hydrolase | | Authors: | Xie, W, Jia, Q. | | Deposit date: | 2021-11-11 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | An engineered PET hydrolase for biodegradation of microplastics in ocean water
To Be Published
|
|
6IOC
 
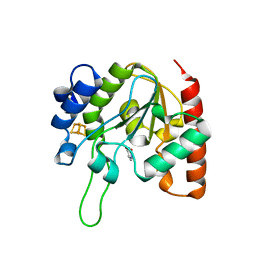 | | The structure of the H109Q mutant of UdgX in complex with uracil | | Descriptor: | IRON/SULFUR CLUSTER, Phage SPO1 DNA polymerase-related protein, URACIL | | Authors: | Xie, W, Tu, J. | | Deposit date: | 2018-10-29 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.624 Å) | | Cite: | Suicide inactivation of the uracil DNA glycosylase UdgX by covalent complex formation.
Nat.Chem.Biol., 15, 2019
|
|
6IOA
 
 | | The structure of UdgX in complex with uracil | | Descriptor: | IRON/SULFUR CLUSTER, Phage SPO1 DNA polymerase-related protein, SULFATE ION, ... | | Authors: | Xie, W, Tu, J. | | Deposit date: | 2018-10-29 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Suicide inactivation of the uracil DNA glycosylase UdgX by covalent complex formation.
Nat.Chem.Biol., 15, 2019
|
|
7W1Q
 
 | |
6IO9
 
 | | The structure of apo-UdgX | | Descriptor: | IRON/SULFUR CLUSTER, Phage SPO1 DNA polymerase-related protein, SULFATE ION | | Authors: | Xie, W, Tu, J. | | Deposit date: | 2018-10-29 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Suicide inactivation of the uracil DNA glycosylase UdgX by covalent complex formation.
Nat.Chem.Biol., 15, 2019
|
|
6IOB
 
 | | The structure of the H109A mutant of UdgX in complex with uracil | | Descriptor: | IRON/SULFUR CLUSTER, Phage SPO1 DNA polymerase-related protein, URACIL | | Authors: | Xie, W, Tu, J. | | Deposit date: | 2018-10-29 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Suicide inactivation of the uracil DNA glycosylase UdgX by covalent complex formation.
Nat.Chem.Biol., 15, 2019
|
|
6L6S
 
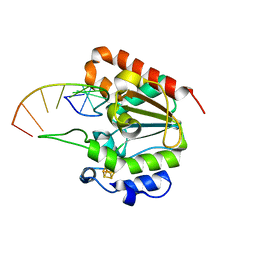 | | The structure of the UdgX mutant H109E crosslinked to single-stranded DNA | | Descriptor: | DNA (5'-D(P*TP*GP*(ORP)P*AP*GP*GP*CP*AP*TP*GP*C)-3'), IRON/SULFUR CLUSTER, Phage SPO1 DNA polymerase-related protein | | Authors: | Xie, W, Tu, J, Zeng, H. | | Deposit date: | 2019-10-29 | | Release date: | 2020-11-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.06425953 Å) | | Cite: | Structural insights into an MsmUdgX mutant capable of both crosslinking and uracil excision capability.
DNA Repair (Amst), 97, 2021
|
|
6KYT
 
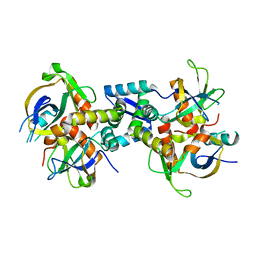 | | The structure of the M. tb toxin MazEF-mt1 complex | | Descriptor: | Antitoxin MazE9, Endoribonuclease MazF9 | | Authors: | Xie, W, Chen, R, Zhou, J. | | Deposit date: | 2019-09-20 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.00101161 Å) | | Cite: | Conserved Conformational Changes in the Regulation ofMycobacterium tuberculosisMazEF-mt1.
Acs Infect Dis., 6, 2020
|
|
6L5B
 
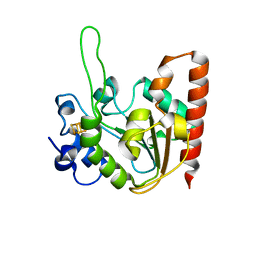 | | The structure of the UdgX mutant H109E at a post-excision state | | Descriptor: | IRON/SULFUR CLUSTER, Uracil DNA glycosylase superfamily protein | | Authors: | Xie, W, Tu, J, Zeng, H. | | Deposit date: | 2019-10-22 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.00004983 Å) | | Cite: | Structural insights into an MsmUdgX mutant capable of both crosslinking and uracil excision capability.
DNA Repair (Amst), 97, 2021
|
|
6IJN
 
 | |
6IER
 
 | | Apo structure of a beta-glucosidase 1317 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, beta-glucosidase 1317 | | Authors: | Xie, W, Liu, X. | | Deposit date: | 2018-09-16 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.246 Å) | | Cite: | Improving the cellobiose-hydrolysis activity and glucose-tolerance of a thermostable beta-glucosidase through rational design.
Int.J.Biol.Macromol., 136, 2019
|
|
6IJM
 
 | |
6IJP
 
 | | The structure of the ADAL-IMP complex | | Descriptor: | Adenosine/AMP deaminase family protein, INOSINIC ACID, ZINC ION | | Authors: | Xie, W, Jia, Q. | | Deposit date: | 2018-10-11 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Alternative conformation induced by substrate binding for Arabidopsis thalianaN6-methyl-AMP deaminase.
Nucleic Acids Res., 47, 2019
|
|
6L94
 
 | | The structure of the dioxygenase ABH1 from mouse | | Descriptor: | FE (II) ION, Nucleic acid dioxygenase ALKBH1 | | Authors: | Xie, W, Wang, C, Li, H, Zhang, Y. | | Deposit date: | 2019-11-08 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.10012341 Å) | | Cite: | ALKBH1 promotes lung cancer by regulating m6A RNA demethylation.
Biochem Pharmacol, 189, 2021
|
|
