7DNC
 
 | | Crystal structure of EV71 3C proteinase in complex with a novel inhibitor | | 分子名称: | 3C protease, ~{N}-[(2~{S})-3-cyclohexyl-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]propan-2-yl]-1~{H}-indole-2-carboxamide | | 著者 | Xie, H, Su, H.X, Li, M.J, Xu, Y.C. | | 登録日 | 2020-12-09 | | 公開日 | 2021-05-05 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.17 Å) | | 主引用文献 | Design, Synthesis, and Biological Evaluation of Peptidomimetic Aldehydes as Broad-Spectrum Inhibitors against Enterovirus and SARS-CoV-2.
J.Med.Chem., 65, 2022
|
|
8H1D
 
 | | Solid-state NMR Structure of Aquaporin Z in its Native Cellular Membranes | | 分子名称: | Aquaporin Z | | 著者 | Xie, H, Zhao, Y, Zhao, W, Chen, Y, Liu, M, Yang, J. | | 登録日 | 2022-10-02 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLID-STATE NMR | | 主引用文献 | Solid-state NMR structure determination of a membrane protein in E. coli cellular inner membrane.
Sci Adv, 9, 2023
|
|
8WE3
 
 | | Crystal structure of human FABP4 complexed with C7 | | 分子名称: | 2-[(3-chloranyl-2-phenyl-phenyl)amino]-5-fluoranyl-benzoic acid, Fatty acid-binding protein, adipocyte | | 著者 | Xie, H, Chen, G.F, Xu, Y.C, Li, M.J. | | 登録日 | 2023-09-16 | | 公開日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Structure-based design of potent FABP4 inhibitors with high selectivity against FABP3.
Eur.J.Med.Chem., 264, 2023
|
|
8WDX
 
 | | Crystal structure of human FABP4 complexed with C3 | | 分子名称: | 1,2-ETHANEDIOL, 2-[(3-chloranyl-2-phenyl-phenyl)amino]-6-methyl-benzoic acid, Fatty acid-binding protein, ... | | 著者 | Xie, H, Chen, G.F, Xu, Y.C. | | 登録日 | 2023-09-16 | | 公開日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structure-based design of potent FABP4 inhibitors with high selectivity against FABP3.
Eur.J.Med.Chem., 264, 2023
|
|
1H6X
 
 | | The role of conserved amino acids in the cleft of the C-terminal family 22 carbohydrate binding module of Clostridium thermocellum Xyn10B in ligand binding | | 分子名称: | CALCIUM ION, ENDO-1,4-BETA-XYLANASE Y | | 著者 | Xie, H, Bolam, D.N, Charnock, S.J, Davies, G.J, Williamson, M.P, Simpson, P.J, Fontes, C.M.G.A, Ferreira, L.M.A, Gilbert, H.J. | | 登録日 | 2001-06-29 | | 公開日 | 2002-06-27 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Clostridium Thermocellum Xyn10B Carbohydrate-Binding Module 22-2: The Role of Conserved Amino Acids in Ligand Binding
Biochemistry, 40, 2001
|
|
1H6Y
 
 | | The role of conserved amino acids in the cleft of the C-terminal family 22 carbohydrate binding module of Clostridium thermocellum Xyn10B in ligand binding | | 分子名称: | CALCIUM ION, ENDO-1,4-BETA-XYLANASE Y | | 著者 | Xie, H, Bolam, D.N, Charnock, S.J, Davies, G.J, Williamson, M.P, Simpson, P.J, Fontes, C.M.G.A, Ferreira, L.M.A, Gilbert, H.J. | | 登録日 | 2001-06-29 | | 公開日 | 2002-06-27 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Clostridium Thermocellum Xyn10B Carbohydrate-Binding Module 22-2: The Role of Conserved Amino Acids in Ligand Binding
Biochemistry, 40, 2001
|
|
2CNC
 
 | | Family 10 xylanase | | 分子名称: | CHLORIDE ION, ENDOXYLANASE, MAGNESIUM ION, ... | | 著者 | Xie, H, Flint, J, Vardakou, M, Lakey, J.H, Lewis, R.J, Gilbert, H.J, Dumon, C. | | 登録日 | 2006-05-19 | | 公開日 | 2006-06-14 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Probing the Structural Basis for the Difference in Thermostability Displayed by Family 10 Xylanases.
J.Mol.Biol., 360, 2006
|
|
7Z6Q
 
 | |
7YF3
 
 | |
7YF4
 
 | |
7Y9C
 
 | |
7YF2
 
 | |
5DJQ
 
 | | The structure of CBB3 cytochrome oxidase. | | 分子名称: | CALCIUM ION, COPPER (II) ION, Cbb3-type cytochrome c oxidase subunit CcoN1, ... | | 著者 | Buschmann, S, Warkentin, E, Xie, H, Kohlstaedt, M, Langer, J.D, Ermler, U, Michel, H. | | 登録日 | 2015-09-02 | | 公開日 | 2016-01-13 | | 最終更新日 | 2016-07-27 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | The structure of cbb3 cytochrome oxidase provides insights into proton pumping.
Science, 329, 2010
|
|
7NB6
 
 | |
7OT9
 
 | |
6Z71
 
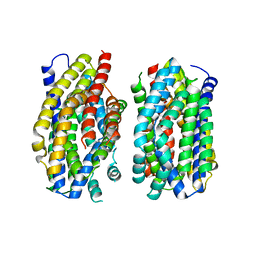 | | Structure of the MATE family multidrug resistance transporter Aq_128 from Aquifex aeolicus in the outward-facing state | | 分子名称: | Aq128 | | 著者 | Zhao, J, Safarian, S, Thielmann, Y, Xie, H, Wang, J, Michel, H. | | 登録日 | 2020-05-29 | | 公開日 | 2021-12-01 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | The structure of the Aquifex aeolicus MATE family multidrug resistance transporter and sequence comparisons suggest the existence of a new subfamily.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6Z70
 
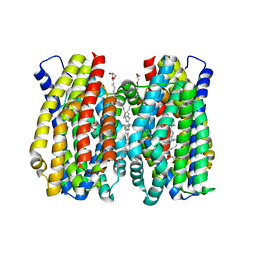 | | Structure of the MATE family multidrug resistance transporter Aq_128 from Aquifex aeolicus in the outward-facing state | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Aq128 | | 著者 | Zhao, J, Safarian, S, Thielmann, Y, Xie, H, Wang, J, Michel, H. | | 登録日 | 2020-05-29 | | 公開日 | 2021-12-01 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The structure of the Aquifex aeolicus MATE family multidrug resistance transporter and sequence comparisons suggest the existence of a new subfamily.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6FV7
 
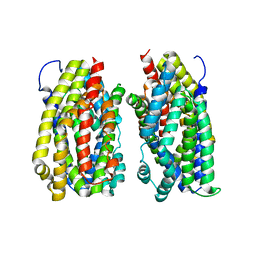 | | Dimer structure of the MATE family multidrug resistance transporter Aq_128 from Aquifex aeolicus in the outward-facing state | | 分子名称: | Aq128 | | 著者 | Zhao, J, Safarian, S, Thielmann, Y, Xie, H, Wang, J, Michel, H. | | 登録日 | 2018-03-01 | | 公開日 | 2019-03-20 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (3.7 Å) | | 主引用文献 | Dimer structure of Aq128 in the outward-facing state
To Be Published
|
|
6FV6
 
 | | Monomer structure of the MATE family multidrug resistance transporter Aq_128 from Aquifex aeolicus in the outward-facing state | | 分子名称: | Aq128 | | 著者 | Zhao, J, Safarian, S, Thielmann, Y, Xie, H, Wang, J, Michel, H. | | 登録日 | 2018-03-01 | | 公開日 | 2019-03-20 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | Monomer structure of Aq128 in the outward-facing state
To Be Published
|
|
6FV8
 
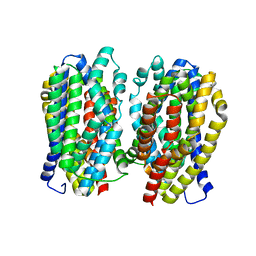 | | Dimer structure of the MATE family multidrug resistance transporter Aq_128 from Aquifex aeolicus in the outward-facing state | | 分子名称: | Aq128 | | 著者 | Zhao, J, Safarian, S, Thielmann, Y, Xie, H, Wang, J, Michel, H. | | 登録日 | 2018-03-01 | | 公開日 | 2019-03-20 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Dimer structure of the MATE family multidrug resistance transporter Aq128
in the outward-facing state
To Be Published
|
|
7TPR
 
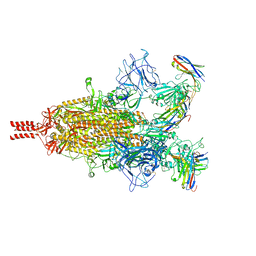 | | Camel nanobodies 7A3 and 8A2 broadly neutralize SARS-CoV-2 variants | | 分子名称: | Nanobody 7A3, Nanobody 8A2, Spike glycoprotein | | 著者 | Butay, K.J, Zhu, J, Dandey, V.P, Hong, J, Kwon, H.J, Chen, C.Z, Duan, Z, Li, D, Ren, H, Liang, T, Martin, N, Esposito, D, Ortega-Rodriguez, U, Xu, M, Xie, H, Ho, M, Cachau, R, Borgnia, M.J. | | 登録日 | 2022-01-25 | | 公開日 | 2022-04-20 | | 実験手法 | ELECTRON MICROSCOPY (2.39 Å) | | 主引用文献 | Camel nanobodies broadly neutralize SARS-CoV-2 variants
bioRxiv, 2021
|
|
7DPV
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with 7-O-methyl-dihydromyricetin | | 分子名称: | (2S,3S)-3,5-dihydroxy-7-methoxy-2-(3,4,5-trihydroxyphenyl)chroman-4-one, 3C-like proteinase | | 著者 | Su, H.X, Zhao, W.F, Xie, H, Li, M.J, Xu, Y.C. | | 登録日 | 2020-12-21 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Identification of pyrogallol as a warhead in design of covalent inhibitors for the SARS-CoV-2 3CL protease.
Nat Commun, 12, 2021
|
|
7DPU
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with 7-O-methyl-myricetin | | 分子名称: | 3C-like proteinase, 7-methoxy-3,5-bis(oxidanyl)-2-[3,4,5-tris(oxidanyl)phenyl]chromen-4-one, GLYCEROL | | 著者 | Su, H.X, Zhao, W.F, Xie, H, Li, M.J, Xu, Y.C. | | 登録日 | 2020-12-21 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Identification of pyrogallol as a warhead in design of covalent inhibitors for the SARS-CoV-2 3CL protease.
Nat Commun, 12, 2021
|
|
7DPP
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with myricetin | | 分子名称: | 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, 3C-like proteinase | | 著者 | Su, H.X, Zhao, W.F, Xie, H, Li, M.J, Xu, Y.C. | | 登録日 | 2020-12-21 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Identification of pyrogallol as a warhead in design of covalent inhibitors for the SARS-CoV-2 3CL protease.
Nat Commun, 12, 2021
|
|
2XFE
 
 | | vCBM60 in complex with galactobiose | | 分子名称: | CALCIUM ION, CARBOHYDRATE BINDING MODULE, beta-D-galactopyranose-(1-4)-beta-D-galactopyranose | | 著者 | Montanier, C, Flint, J.E, Bolam, D.N, Xie, H, Liu, Z, Rogowski, A, Weiner, D.P, Nurizzo, D, Roberts, S.M, Turkenburg, J.P, Davies, G.J, Gilbert, H.J. | | 登録日 | 2010-05-21 | | 公開日 | 2010-06-16 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Circular Permutation Provides an Evolutionary Link between Two Families of Calcium-Dependent Carbohydrate Binding Modules.
J.Biol.Chem., 285, 2010
|
|
