7XLC
 
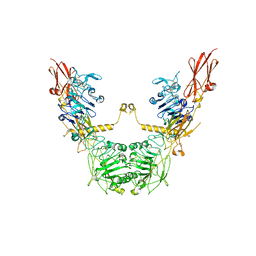 | | conformation II of apo-IGF1R states | | Descriptor: | Insulin-like growth factor 1 receptor | | Authors: | Xi, Z, Cang, W. | | Deposit date: | 2022-04-21 | | Release date: | 2023-04-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | conformation II of apo-IGF1R states
To Be Published
|
|
2LQ9
 
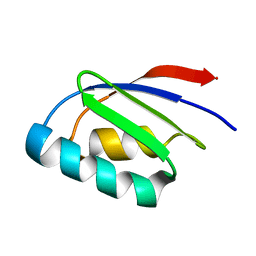 | |
2LW3
 
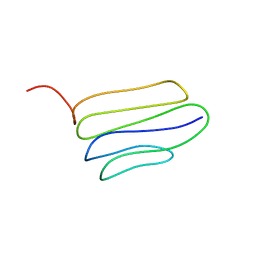 | | Solution structure of the soluble domain of MmpS4 from Mycobacterium tuberculosis | | Descriptor: | Putative membrane protein mmpS4 | | Authors: | Xi, Z, Sun, P, Wang, W, Lai, C, Wu, F, Tian, C. | | Deposit date: | 2012-07-19 | | Release date: | 2013-03-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Discovery of a Siderophore Export System Essential for Virulence of Mycobacterium tuberculosis
Plos Pathog., 9, 2013
|
|
7YRR
 
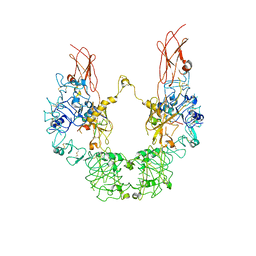 | | Cryo-EM structure of IGF1R with two IGF1 complex | | Descriptor: | Insulin-like growth factor 1 receptor, Isoform 3 of Insulin-like growth factor I | | Authors: | Xi, Z, Cang, W. | | Deposit date: | 2022-08-10 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structure of IGF1R with two IGF1 complex at 4.3 angstroms resolution
To Be Published
|
|
8IJ5
 
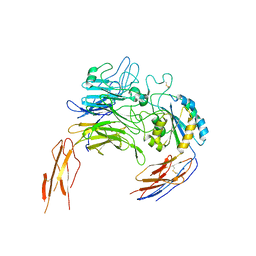 | |
7VT0
 
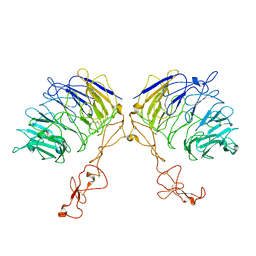 | | Dimer structure of SORLA | | Descriptor: | Sortilin-related receptor | | Authors: | Xi, Z, Cang, W, Chuang, L. | | Deposit date: | 2021-10-27 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures reveal distinct apo conformations of sortilin-related receptor SORLA.
Biochem.Biophys.Res.Commun., 600, 2022
|
|
1MP7
 
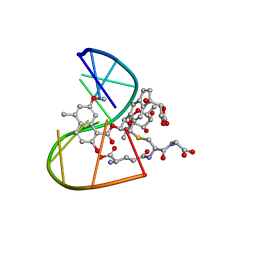 | | A Third Complex of Post-Activated Neocarzinostatin Chromophore with DNA. Bulge DNA Binding from the Minor Groove | | Descriptor: | 5'-D(*GP*CP*CP*AP*GP*AP*GP*AP*GP*C)-3', [(R)-4-((1,3-DIOXOLANE-2-OXY)-4-(S)-YL)-4-HYDROXY]-(R)-10-(2-METHYLAMINO-5-METHYL-2,6-DIDEOXYGALACTOPYRANOSYL-OXY)-(R)-11-(2-HYDROXY-5-METHYL-7-METHOXY-1-NAPHTHOYL-OXY)-(R)-12-S-GLUTATHIONYL-4,10,11,12-TETRAHYDROINDACENE | | Authors: | Kwon, Y, Xi, Z, Kappen, L.S, Goldberg, I.H, Gao, X. | | Deposit date: | 2002-09-11 | | Release date: | 2003-02-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | New Complex of Post-Activated Neocarzinostatin Chromophore with DNA: Bulge DNA Binding from the Minor Groove
Biochemistry, 42, 2003
|
|
7QA9
 
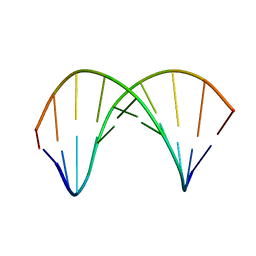 | | 10bp DNA/DNA duplex | | Descriptor: | DNA (5'-D(*CP*CP*AP*TP*TP*AP*TP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*AP*TP*AP*AP*TP*GP*G)-3') | | Authors: | Li, Q, Trajkovski, M, Fan, C, Chen, J, Zhou, Y, Lu, K, Li, H, Su, X, Xi, Z, Plavec, J, Zhou, C. | | Deposit date: | 2021-11-16 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | 4'-SCF 3 -Labeling Constitutes a Sensitive 19 F NMR Probe for Characterization of Interactions in the Minor Groove of DNA.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
6LPI
 
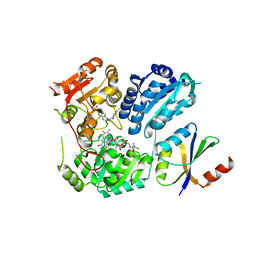 | | Crystal Structure of AHAS holo-enzyme | | Descriptor: | Acetolactate synthase isozyme 1 large subunit, Acetolactate synthase isozyme 1 small subunit, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Zhang, Y, Yang, X, Xi, Z, Shen, Y. | | Deposit date: | 2020-01-10 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.849 Å) | | Cite: | Molecular architecture of the acetohydroxyacid synthase holoenzyme.
Biochem.J., 477, 2020
|
|
6A9D
 
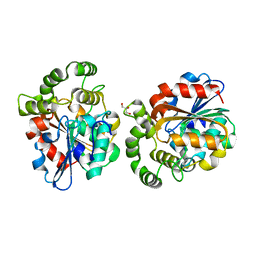 | |
7W0V
 
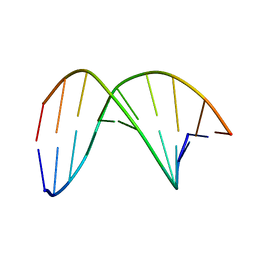 | | C4'-SCF3-DT modifeid DNA-DNA duplex | | Descriptor: | DNA (5'-D(*CP*CP*AP*TP*(DSW)P*AP*TP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*AP*TP*AP*AP*TP*GP*G)-3') | | Authors: | Li, Q, Trajkovski, M, Fan, C, Chen, J, Zhou, Y, Lu, K, Li, H, Su, X, Xi, Z, Plavec, J, Zhou, C. | | Deposit date: | 2021-11-18 | | Release date: | 2022-11-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 4'-SCF 3 -Labeling Constitutes a Sensitive 19 F NMR Probe for Characterization of Interactions in the Minor Groove of DNA.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
6J2R
 
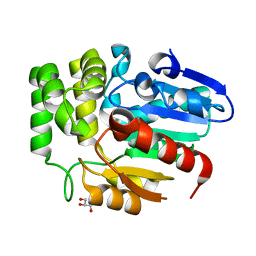 | | Crystal structure of Striga hermonthica HTL8 (ShHTL8) | | Descriptor: | GLYCEROL, Hyposensitive to light 8 | | Authors: | Zhang, Y.Y, Xi, Z. | | Deposit date: | 2019-01-02 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure and biochemical characterization of Striga hermonthica HYPO-SENSITIVE TO LIGHT 8 (ShHTL8) in strigolactone signaling pathway.
Biochem.Biophys.Res.Commun., 523, 2020
|
|
