5X5B
 
 | | Prefusion structure of SARS-CoV spike glycoprotein, conformation 2 | | Descriptor: | Spike glycoprotein | | Authors: | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-03 | | Last modified: | 2017-05-24 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
5X59
 
 | | Prefusion structure of MERS-CoV spike glycoprotein, three-fold symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S protein | | Authors: | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-03 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
5X5C
 
 | | Prefusion structure of MERS-CoV spike glycoprotein, conformation 1 | | Descriptor: | S protein | | Authors: | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-03 | | Last modified: | 2017-05-24 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
6KXT
 
 | | BON1-C2B | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Protein BONZAI 1, SULFATE ION | | Authors: | Wang, Q.C, Jiang, M.Q, Isupov, M.N, Sun, L.F, Wu, Y.K. | | Deposit date: | 2019-09-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal Structure of an Arabidopsis Copine providing insights into this protein family
To be published
|
|
5X5F
 
 | | Prefusion structure of MERS-CoV spike glycoprotein, conformation 2 | | Descriptor: | S protein | | Authors: | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-03 | | Last modified: | 2017-05-24 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
6LUO
 
 | | Structure of nurse shark beta-2-microglobulin | | Descriptor: | Beta-2-microglobulin | | Authors: | Xia, C, Wu, Y. | | Deposit date: | 2020-01-30 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | The Structure of a Peptide-Loaded Shark MHC Class I Molecule Reveals Features of the Binding between beta 2 -Microglobulin and H Chain Conserved in Evolution.
J Immunol., 2021
|
|
6MHN
 
 | | Photoactive Yellow Protein with covalently bound 3-chloro-4-hydroxycinnamic acid chromophore | | Descriptor: | (2E)-3-(3-chloro-4-hydroxyphenyl)prop-2-enoic acid, Photoactive yellow protein | | Authors: | Thomson, B.D, Both, J, Wu, Y, Parrish, R.M, Martinez, T, Boxer, S.G. | | Deposit date: | 2018-09-18 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Perturbation of Short Hydrogen Bonds in Photoactive Yellow Protein via Noncanonical Amino Acid Incorporation.
J.Phys.Chem.B, 123, 2019
|
|
6MKT
 
 | | Photoactive Yellow Protein with 3-chlorotyrosine substituted at position 42 | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Thomson, B.D, Both, J, Wu, Y, Parrish, R.M, Martinez, T, Boxer, S.G. | | Deposit date: | 2018-09-26 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Perturbation of Short Hydrogen Bonds in Photoactive Yellow Protein via Noncanonical Amino Acid Incorporation.
J.Phys.Chem.B, 123, 2019
|
|
6MMD
 
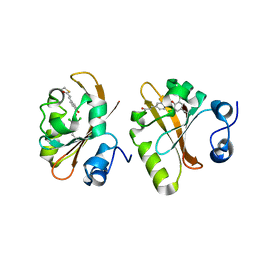 | | Photoactive Yellow Protein with 3,5-dichlorotyrosine substituted at position 42 | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Thomson, B.D, Both, J, Wu, Y, Parrish, R.M, Martinez, T, Boxer, S.G. | | Deposit date: | 2018-09-30 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.228 Å) | | Cite: | Perturbation of Short Hydrogen Bonds in Photoactive Yellow Protein via Noncanonical Amino Acid Incorporation.
J.Phys.Chem.B, 123, 2019
|
|
5V2G
 
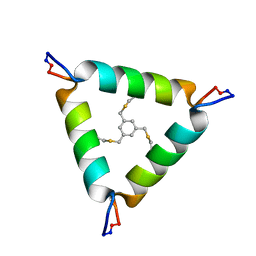 | | De Novo Design of Novel Covalent Constrained Meso-size Peptide Scaffolds with Unique Tertiary Structures | | Descriptor: | 1,3,5-tris(bromomethyl)benzene, 20-mer Peptide | | Authors: | Dang, B, Wu, H, Mulligan, V.K, Mravic, M, Wu, Y, Lemmin, T, Ford, A, Silva, D, Baker, D, DeGrado, W.F. | | Deposit date: | 2017-03-03 | | Release date: | 2017-09-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | De novo design of covalently constrained mesosize protein scaffolds with unique tertiary structures.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6O3P
 
 | |
6OIX
 
 | | Structure of Escherichia coli dGTPase bound to GTP | | Descriptor: | Deoxyguanosinetriphosphate triphosphohydrolase, GUANOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION | | Authors: | Barnes, C.O, Wu, Y, Calero, G. | | Deposit date: | 2019-04-09 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | The crystal structure of dGTPase reveals the molecular basis of dGTP selectivity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OIY
 
 | | Structure of Escherichia coli bound to dGTP | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, Deoxyguanosinetriphosphate triphosphohydrolase, MANGANESE (II) ION | | Authors: | Barnes, C.O, Wu, Y, Calero, G. | | Deposit date: | 2019-04-09 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | The crystal structure of dGTPase reveals the molecular basis of dGTP selectivity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6ANF
 
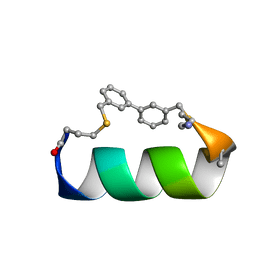 | | Design of a short thermo-stable alpha-helix embedded in a macrocycle | | Descriptor: | 3,3'-dimethyl-1,1'-biphenyl, Capped-strapped peptide | | Authors: | Wu, H, Acharyya, A, Wu, Y, Liu, L, Jo, H, Gai, F, DeGrado, W.F. | | Deposit date: | 2017-08-13 | | Release date: | 2018-02-21 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Design of a Short Thermally Stable alpha-Helix Embedded in a Macrocycle.
Chembiochem, 19, 2018
|
|
5WOD
 
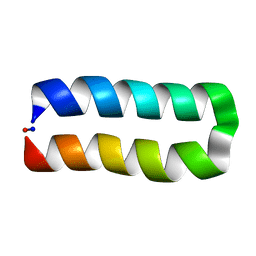 | |
5ZL2
 
 | |
5ZKX
 
 | |
5WSH
 
 | | Structure of HLA-A2 P130 | | Descriptor: | Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, GLY-VAL-TRP-ILE-ARG-THR-PRO-THR-ALA, ... | | Authors: | Zhang, Y, Wu, Y, Qi, J, Liu, J, Gao, G.F, Meng, S. | | Deposit date: | 2016-12-07 | | Release date: | 2017-12-20 | | Last modified: | 2019-01-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CD8+T-Cell Response-Associated Evolution of Hepatitis B Virus Core Protein and Disease Progress.
J. Virol., 92, 2018
|
|
5XMM
 
 | | FLA-E*01801-167W/S | | Descriptor: | Beta-2-microglobulin, Gag polyprotein, MHC class I antigen alpha chain | | Authors: | Liang, R, Sun, Y, Wang, J, Wu, Y, Zhang, N, Xia, C. | | Deposit date: | 2017-05-15 | | Release date: | 2017-12-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Major Histocompatibility Complex Class I (FLA-E*01801) Molecular Structure in Domestic Cats Demonstrates Species-Specific Characteristics in Presenting Viral Antigen Peptides
J. Virol., 92, 2018
|
|
5XMF
 
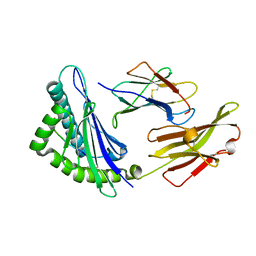 | | Crystal structure of feline MHC class I for 2,1 angstrom | | Descriptor: | Beta-2-microglobulin, Gag polyprotein, MHC class I antigen alpha chain | | Authors: | Liang, R, Sun, Y, Wang, J, Wu, Y, Zhang, N, Xia, C. | | Deposit date: | 2017-05-15 | | Release date: | 2017-12-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Major Histocompatibility Complex Class I (FLA-E*01801) Molecular Structure in Domestic Cats Demonstrates Species-Specific Characteristics in Presenting Viral Antigen Peptides
J. Virol., 92, 2018
|
|
2JR6
 
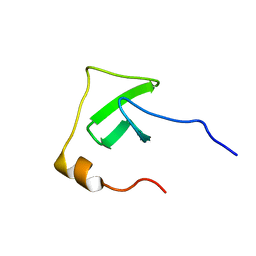 | | Solution structure of UPF0434 protein NMA0874. Northeast Structural Genomics Target MR32 | | Descriptor: | UPF0434 protein NMA0874 | | Authors: | Ghosh, A, Singarapu, K.K, Wu, Y, Liu, G, Sukumaran, D, Chen, C.X, Nwosu, C, Owens, L, Xiao, R, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-20 | | Release date: | 2007-07-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of UPF0434 protein NMA0874.
To be Published
|
|
2JS3
 
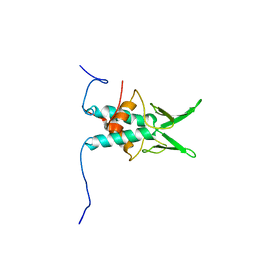 | | NMR Structure of protein Q6N9A4_RHOPA. Northeast Structural Genomics Consortium target RpT8 | | Descriptor: | Uncharacterized protein | | Authors: | Singarapu, K.K, Wu, Y, Yee, A, Eletsky, A, Sukumaran, D, Garcia, M, Xiao, R, Bansal, S, Baran, M.C, Montelione, G.T, James, P.H, Arrowsmith, C.H, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-29 | | Release date: | 2007-07-31 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR structure of protein Q6N9A4_RHOPA.
To be Published
|
|
2KI8
 
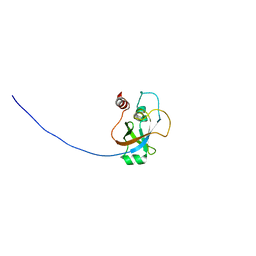 | | Solution NMR structure of tungsten formylmethanofuran dehydrogenase subunit D from Archaeoglobus fulgidus, Northeast Structural Genomics Consortium target AtT7 | | Descriptor: | Tungsten formylmethanofuran dehydrogenase, subunit D (FwdD-2) | | Authors: | Eletsky, A, Wu, Y, Yee, A, Fares, C, Lee, H.W, Arrowsmith, C.H, Prestegard, J.H, Szyperski, T, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-04-28 | | Release date: | 2009-05-26 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of tungsten formylmethanofuran dehydrogenase subunit D from Archaeoglobus fulgidus
To be Published
|
|
2KKV
 
 | | Solution NMR structure of an integrase domain from protein SPA4288 from Salmonella enterica, Northeast Structural Genomics Consortium Target SlR105H | | Descriptor: | Integrase | | Authors: | Mills, J.L, Wu, Y, Sukumaran, D.K, Belote, R.L, Ciccosanti, C, Jiang, M, Nair, R, Rost, B, Acton, T.B, Xiao, R, Swapna, G.V.T, Everett, J.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-29 | | Release date: | 2009-08-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of an integrase domain from protein SPA4288 from Salmonella enterica, Northeast Structural Genomics Consortium Target SlR105H
To be Published
|
|
2KZ2
 
 | | Calmodulin, C-terminal domain, F92E mutant | | Descriptor: | CALCIUM ION, Calmodulin | | Authors: | Korendovych, I, Kulp, D, Wu, Y, Cheng, H, Roder, H, DeGrado, W. | | Deposit date: | 2010-06-10 | | Release date: | 2011-04-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Design of a switchable eliminase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
