6UI7
 
 | | HBV T=4 149C3A | | Descriptor: | Core protein | | Authors: | Wu, W, Watts, N.R, Cheng, N, Huang, R, Steven, A, Wingfield, P.T. | | Deposit date: | 2019-09-30 | | Release date: | 2019-11-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Expression of quasi-equivalence and capsid dimorphism in the Hepadnaviridae.
Plos Comput.Biol., 16, 2020
|
|
6UI6
 
 | | HBV T=3 149C3A | | Descriptor: | Core protein | | Authors: | Wu, W, Watts, N.R, Cheng, N, Huang, R, Steven, A, Wingfield, P.T. | | Deposit date: | 2019-09-30 | | Release date: | 2019-11-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Expression of quasi-equivalence and capsid dimorphism in the Hepadnaviridae.
Plos Comput.Biol., 16, 2020
|
|
3Q9L
 
 | | The structure of the dimeric E.coli MinD-ATP complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Septum site-determining protein minD | | Authors: | Wu, W, Park, K.-T, Lutkenhaus, J, Holyoak, T. | | Deposit date: | 2011-01-08 | | Release date: | 2011-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.343 Å) | | Cite: | Determination of the structure of the MinD-ATP complex reveals the orientation of MinD on the membrane and the relative location of the binding sites for MinE and MinC.
Mol.Microbiol., 79, 2011
|
|
6LB8
 
 | | Crystal structure of the Ca2+-free T4L-MICU1-MICU2 complex | | Descriptor: | Calcium uptake protein 2, mitochondrial, Endolysin,Calcium uptake protein 1 | | Authors: | Wu, W, Shen, Q, Zheng, J, Jia, Z. | | Deposit date: | 2019-11-13 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.283 Å) | | Cite: | The structure of the MICU1-MICU2 complex unveils the regulation of the mitochondrial calcium uniporter.
Embo J., 39, 2020
|
|
6LB7
 
 | | Crystal structure of the Ca2+-free and Ca2+-bound MICU1-MICU2 complex | | Descriptor: | CALCIUM ION, Calcium uptake protein 1, mitochondrial, ... | | Authors: | Wu, W, Shen, Q, Zheng, J, Jia, Z. | | Deposit date: | 2019-11-13 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | The structure of the MICU1-MICU2 complex unveils the regulation of the mitochondrial calcium uniporter.
Embo J., 39, 2020
|
|
2M2U
 
 | |
2M2W
 
 | | Ternary complex of ASFV Pol X with DNA and MgdGTP | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(P*GP*GP*CP*GP*AP*AP*GP*CP*CP*GP*GP*GP*TP*GP*CP*GP*AP*AP*GP*CP*AP*CP*(DOC))-3', MAGNESIUM ION, ... | | Authors: | Wu, W, Su, M, Tsai, M. | | Deposit date: | 2013-01-03 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | How a low-fidelity DNA polymerase chooses non-Watson-Crick from Watson-Crick incorporation.
J.Am.Chem.Soc., 136, 2014
|
|
2M2T
 
 | | ASFV Pol X structure | | Descriptor: | Repair DNA polymerase X | | Authors: | Wu, W, Su, M, Tsai, M. | | Deposit date: | 2013-01-03 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | How a low-fidelity DNA polymerase chooses non-Watson-Crick from Watson-Crick incorporation.
J.Am.Chem.Soc., 136, 2014
|
|
2M2V
 
 | |
6INB
 
 | |
6INC
 
 | | Crystal structure of an acetolactate decarboxylase from Klebsiella pneumoniae | | Descriptor: | 1,2-ETHANEDIOL, Alpha-acetolactate decarboxylase, CHLORIDE ION, ... | | Authors: | Wu, W, Zhang, Q, Bartlam, M. | | Deposit date: | 2018-10-24 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | Structural characterization of an acetolactate decarboxylase from Klebsiella pneumoniae
Biochem. Biophys. Res. Commun., 509, 2019
|
|
6JC4
 
 | |
7FJ8
 
 | |
7FJ9
 
 | | KpAckA (PduW) with AMPPNP complex structure | | Descriptor: | ACETATE ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Probable propionate kinase | | Authors: | Wu, W, Zhang, Q, Bartlam, M. | | Deposit date: | 2021-08-03 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | KpAckA (PduW) with AMPPNP complex structure
To Be Published
|
|
7FJ7
 
 | |
7FJA
 
 | | KpAckA (PduW) with AMPPNP, ethylene glycol complex structure | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Probable propionate kinase | | Authors: | Wu, W, Zhang, Q, Bartlam, M. | | Deposit date: | 2021-08-03 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | KpAckA (PduW) with AMPPNP, ethylene glycol complex structure
To Be Published
|
|
3ZL9
 
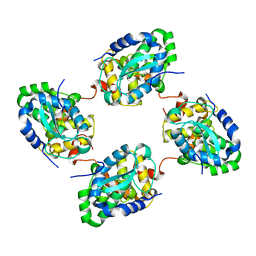 | | Crystal structure of the nucleocapsid protein from Schmallenberg virus | | Descriptor: | NUCLEOCAPSID PROTEIN | | Authors: | Ariza, A, Tanner, S.J, Walter, C.T, Dent, K.C, Shepherd, D.A, Wu, W, Matthews, S.V, Hiscox, J.A, Green, T.J, Luo, M, Elliot, R.M, Ashcroft, A.E, Stonehouse, N.J, Ranson, N.A, Barr, J.N, Edwards, T.A. | | Deposit date: | 2013-01-29 | | Release date: | 2013-05-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Nucleocapsid Protein Structures from Orthobunyaviruses Reveal Insight Into Ribonucleoprotein Architecture and RNA Polymerization.
Nucleic Acids Res., 41, 2013
|
|
3ZLA
 
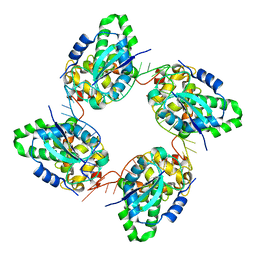 | | Crystal structure of the nucleocapsid protein from Bunyamwera virus bound to RNA | | Descriptor: | NUCLEOPROTEIN, RNA | | Authors: | Ariza, A, Tanner, S.J, Walter, C.T, Dent, K.C, Shepherd, D.A, Wu, W, Matthews, S.V, Hiscox, J.A, Green, T.J, Luo, M, Elliot, R.M, Ashcroft, A.E, Stonehouse, N.J, Ranson, N.A, Barr, J.N, Edwards, T.A. | | Deposit date: | 2013-01-29 | | Release date: | 2013-05-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Nucleocapsid Protein Structures from Orthobunyaviruses Reveal Insight Into Ribonucleoprotein Architecture and RNA Polymerization.
Nucleic Acids Res., 41, 2013
|
|
6G7L
 
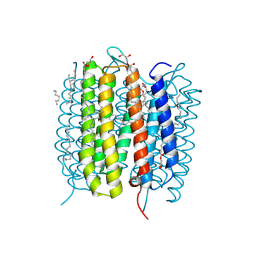 | | Retinal isomerization in bacteriorhodopsin revealed by a femtosecond X-ray laser: 8.3 ms state structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, ... | | Authors: | Nogly, P, Weinert, T, James, D, Cabajo, S, Ozerov, D, Furrer, A, Gashi, D, Borin, V, Skopintsev, P, Jaeger, K, Nass, K, Bath, P, Bosman, R, Koglin, J, Seaberg, M, Lane, T, Kekilli, D, Bruenle, S, Tanaka, T, Wu, W, Milne, C, White, T, Barty, A, Weierstall, U, Panneels, V, Nango, E, Iwata, S, Hunter, M, Schapiro, I, Schertler, G, Neutze, R, Standfuss, J. | | Deposit date: | 2018-04-06 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Retinal isomerization in bacteriorhodopsin captured by a femtosecond x-ray laser.
Science, 361, 2018
|
|
6G7H
 
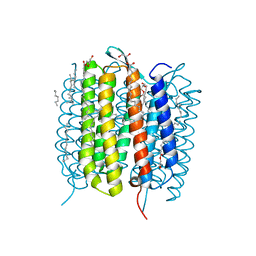 | | Retinal isomerization in bacteriorhodopsin revealed by a femtosecond X-ray laser: resting state structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, ... | | Authors: | Nogly, P, Weinert, T, James, D, Cabajo, S, Ozerov, D, Furrer, A, Gashi, D, Borin, V, Skopintsev, P, Jaeger, K, Nass, K, Bath, P, Bosman, R, Koglin, J, Seaberg, M, Lane, T, Kekilli, D, Bruenle, S, Tanaka, T, Wu, W, Milne, C, White, T, Barty, A, Weierstall, U, Panneels, V, Nango, E, Iwata, S, Hunter, M, Schapiro, I, Schertler, G, Neutze, R, Standfuss, J. | | Deposit date: | 2018-04-06 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Retinal isomerization in bacteriorhodopsin captured by a femtosecond x-ray laser.
Science, 361, 2018
|
|
6G7I
 
 | | Retinal isomerization in bacteriorhodopsin revealed by a femtosecond X-ray laser: 49-406 fs state structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, ... | | Authors: | Nogly, P, Weinert, T, James, D, Cabajo, S, Ozerov, D, Furrer, A, Gashi, D, Borin, V, Skopintsev, P, Jaeger, K, Nass, K, Bath, P, Bosman, R, Koglin, J, Seaberg, M, Lane, T, Kekilli, D, Bruenle, S, Tanaka, T, Wu, W, Milne, C, White, T, Barty, A, Weierstall, U, Panneels, V, Nango, E, Iwata, S, Hunter, M, Schapiro, I, Schertler, G, Neutze, R, Standfuss, J. | | Deposit date: | 2018-04-06 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Retinal isomerization in bacteriorhodopsin captured by a femtosecond x-ray laser.
Science, 361, 2018
|
|
6G7J
 
 | | Retinal isomerization in bacteriorhodopsin revealed by a femtosecond X-ray laser: 457-646 fs state structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, ... | | Authors: | Nogly, P, Weinert, T, James, D, Cabajo, S, Ozerov, D, Furrer, A, Gashi, D, Borin, V, Skopintsev, P, Jaeger, K, Nass, K, Bath, P, Bosman, R, Koglin, J, Seaberg, M, Lane, T, Kekilli, D, Bruenle, S, Tanaka, T, Wu, W, Milne, C, White, T, Barty, A, Weierstall, U, Panneels, V, Nango, E, Iwata, S, Hunter, M, Schapiro, I, Schertler, G, Neutze, R, Standfuss, J. | | Deposit date: | 2018-04-06 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Retinal isomerization in bacteriorhodopsin captured by a femtosecond x-ray laser.
Science, 361, 2018
|
|
6G7K
 
 | | Retinal isomerization in bacteriorhodopsin revealed by a femtosecond X-ray laser: 10 ps state structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, ... | | Authors: | Nogly, P, Weinert, T, James, D, Cabajo, S, Ozerov, D, Furrer, A, Gashi, D, Borin, V, Skopintsev, P, Jaeger, K, Nass, K, Bath, P, Bosman, R, Koglin, J, Seaberg, M, Lane, T, Kekilli, D, Bruenle, S, Tanaka, T, Wu, W, Milne, C, White, T, Barty, A, Weierstall, U, Panneels, V, Nango, E, Iwata, S, Hunter, M, Schapiro, I, Schertler, G, Neutze, R, Standfuss, J. | | Deposit date: | 2018-04-06 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Retinal isomerization in bacteriorhodopsin captured by a femtosecond x-ray laser.
Science, 361, 2018
|
|
2XYY
 
 | | De Novo model of Bacteriophage P22 procapsid coat protein | | Descriptor: | COAT PROTEIN | | Authors: | Chen, D.-H, Baker, M.L, Hryc, C.F, DiMaio, F, Jakana, J, Wu, W, Dougherty, M, Haase-Pettingell, C, Schmid, M.F, Jiang, W, Baker, D, King, J.A, Chiu, W. | | Deposit date: | 2010-11-19 | | Release date: | 2011-02-02 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural Basis for Scaffolding-Mediated Assembly and Maturation of a DsDNA Virus.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2XYZ
 
 | | De Novo model of Bacteriophage P22 virion coat protein | | Descriptor: | COAT PROTEIN | | Authors: | Chen, D.-H, Baker, M.L, Hryc, C.F, DiMaio, F, Jakana, J, Wu, W, Dougherty, M, Haase-Pettingell, C, Schmid, M.F, Jiang, W, Baker, D, King, J.A, Chiu, W. | | Deposit date: | 2010-11-19 | | Release date: | 2011-02-02 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural Basis for Scaffolding-Mediated Assembly and Maturation of a DsDNA Virus.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
