3V1Q
 
 | |
8ECE
 
 | | E. coli L-asparaginase II mutant (V27T) in complex with L-Glu | | Descriptor: | 1,2-ETHANEDIOL, GLUTAMIC ACID, L-asparaginase 2 | | Authors: | Strzelczyk, P, Wlodawer, A, Lubkowski, J. | | Deposit date: | 2022-09-01 | | Release date: | 2022-11-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The E. coli L-asparaginase V27T mutant: structural and functional characterization and comparison with theoretical predictions.
Febs Lett., 596, 2022
|
|
8ECD
 
 | | E. coli L-asparaginase II mutant (V27T) in complex with L-Asp | | Descriptor: | 1,2-ETHANEDIOL, ASPARTIC ACID, CITRIC ACID, ... | | Authors: | Strzelczyk, P, Wlodawer, A, Lubkowski, J. | | Deposit date: | 2022-09-01 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | The E. coli L-asparaginase V27T mutant: structural and functional characterization and comparison with theoretical predictions.
Febs Lett., 596, 2022
|
|
5DZK
 
 | | Crystal structure of the active form of the proteolytic complex clpP1 and clpP2 | | Descriptor: | ATP-dependent Clp protease proteolytic subunit 1, ATP-dependent Clp protease proteolytic subunit 2, BEZ-LEU-LEU | | Authors: | LI, M, Wlodawer, A, Maurizi, M. | | Deposit date: | 2015-09-25 | | Release date: | 2016-02-17 | | Last modified: | 2016-04-13 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Structure and Functional Properties of the Active Form of the Proteolytic Complex, ClpP1P2, from Mycobacterium tuberculosis.
J.Biol.Chem., 291, 2016
|
|
5N0H
 
 | | Crystal structure of NDM-1 in complex with hydrolyzed meropenem - new refinement | | Descriptor: | (2S,3R)-2-[(2S,3R)-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfan yl-3-methyl-2,3-dihydro-1H-pyrrole-5-carboxylic acid, GLYCEROL, Metallo-beta-lactamase type 2, ... | | Authors: | Raczynska, J.E, Shabalin, I.G, Jaskolski, M, Minor, W, Wlodawer, A, King, D.T, Strynadka, N.C.J. | | Deposit date: | 2017-02-03 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
5N0I
 
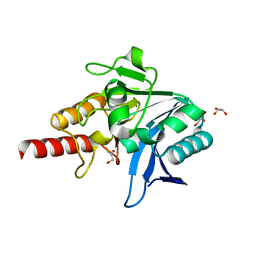 | | Crystal structure of NDM-1 in complex with beta-mercaptoethanol - new refinement | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Raczynska, J.E, Shabalin, I.G, Jaskolski, M, Minor, W, Wlodawer, A, King, D.T, Strynadka, N.C.J. | | Deposit date: | 2017-02-03 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
5NBK
 
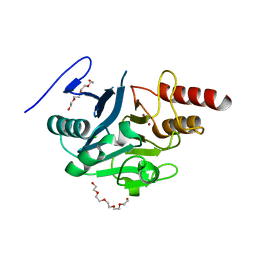 | | NDM-1 metallo-beta-lactamase: a parsimonious interpretation of the diffraction data | | Descriptor: | CHLORIDE ION, HEXAETHYLENE GLYCOL, Metallo-beta-lactamase type 2, ... | | Authors: | Raczynska, J.E, Shabalin, I.G, Jaskolski, M, Minor, W, Wlodawer, A. | | Deposit date: | 2017-03-02 | | Release date: | 2018-10-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
5DUY
 
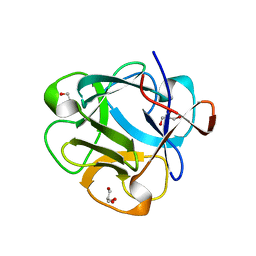 | | Structure of lectin from the sea mussel Crenomytilus grayanus | | Descriptor: | GLYCEROL, GalNAc/Gal-specific lectin | | Authors: | Lubkowski, J, Jakob, M, O'Keefe, B, Wlodawer, A. | | Deposit date: | 2015-09-21 | | Release date: | 2015-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure of a lectin from the sea mussel Crenomytilus grayanus (CGL).
Acta Crystallogr.,Sect.F, 71, 2015
|
|
8R2C
 
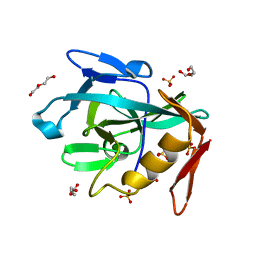 | | Crystal structure of the Vint domain from Tetrahymena thermophila | | Descriptor: | DI(HYDROXYETHYL)ETHER, SULFATE ION, von willebrand factor type A (VWA) domain was originally protein | | Authors: | Iwai, H, Beyer, H.M, Johannson, J.E, Li, M, Wlodawer, A. | | Deposit date: | 2023-11-03 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The three-dimensional structure of the Vint domain from Tetrahymena thermophila suggests a ligand-regulated cleavage mechanism by the HINT fold.
Febs Lett., 598, 2024
|
|
4RLD
 
 | |
1BTI
 
 | | CREVICE-FORMING MUTANTS IN THE RIGID CORE OF BOVINE PANCREATIC TRYPSIN INHIBITOR: CRYSTAL STRUCTURES OF F22A, Y23A, N43G, AND F45A | | Descriptor: | BOVINE PANCREATIC TRYPSIN INHIBITOR | | Authors: | Housset, D, Tao, F, Kim, K.-S, Fuchs, J, Woodward, C, Wlodawer, A. | | Deposit date: | 1991-07-11 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crevice-forming mutants in the rigid core of bovine pancreatic trypsin inhibitor: crystal structures of F22A, Y23A, N43G, and F45A.
Protein Sci., 2, 1993
|
|
1BPT
 
 | | CREVICE-FORMING MUTANTS OF BPTI: CRYSTAL STRUCTURES OF F22A, Y23A, N43G, AND F45A | | Descriptor: | BOVINE PANCREATIC TRYPSIN INHIBITOR, PHOSPHATE ION | | Authors: | Housset, D, Wlodawer, A, Tao, F, Fuchs, J, Woodward, C. | | Deposit date: | 1991-12-11 | | Release date: | 1993-01-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crevice-forming mutants in the rigid core of bovine pancreatic trypsin inhibitor: crystal structures of F22A, Y23A, N43G, and F45A.
Protein Sci., 2, 1993
|
|
3V1O
 
 | |
4MON
 
 | | ORTHORHOMBIC MONELLIN | | Descriptor: | MONELLIN | | Authors: | Bujacz, G, Wlodawer, A. | | Deposit date: | 1997-03-04 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of monellin refined to 2.3 a resolution in the orthorhombic crystal form.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
7R6A
 
 | |
7R69
 
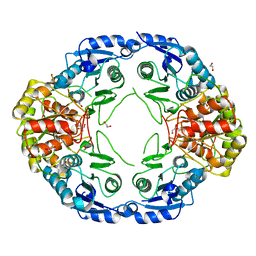 | |
7R6B
 
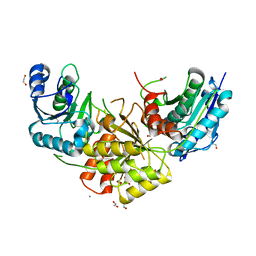 | | Crystal structure of mutant R43D/L124D/R125A/C273S of L-Asparaginase I from Yersinia pestis | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, L-asparaginase I | | Authors: | Strzelczyk, P, Wlodawer, A, Lubkowski, J. | | Deposit date: | 2021-06-22 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The dimeric form of bacterial l-asparaginase YpAI is fully active.
Febs J., 290, 2023
|
|
4O1S
 
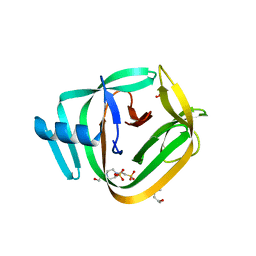 | | Crystal structure of TvoVMA intein | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, SULFATE ION, ... | | Authors: | Aranko, A.S, Oeemig, J.S, Zhou, D, Kajander, T, Wlodawer, A, Iwai, H. | | Deposit date: | 2013-12-16 | | Release date: | 2014-03-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7008 Å) | | Cite: | Structure-based engineering and comparison of novel split inteins for protein ligation.
Mol Biosyst, 10, 2014
|
|
4O1R
 
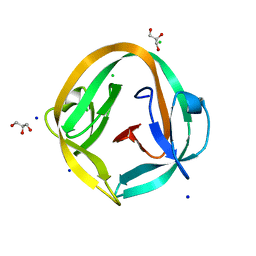 | | Crystal structure of NpuDnaB intein | | Descriptor: | CHLORIDE ION, GLYCEROL, Replicative DNA helicase, ... | | Authors: | Aranko, A.S, Oeemig, J.S, Zhou, D, Kajander, T, Wlodawer, A, Iwai, H. | | Deposit date: | 2013-12-16 | | Release date: | 2014-03-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-based engineering and comparison of novel split inteins for protein ligation.
Mol Biosyst, 10, 2014
|
|
3V1R
 
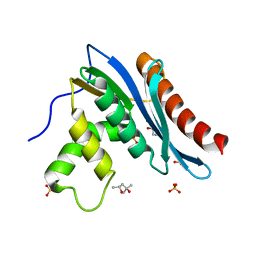 | | Crystal structures of the reverse transcriptase-associated ribonuclease H domain of XMRV with inhibitor beta-thujaplicinol | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2,7-dihydroxy-4-(propan-2-yl)cyclohepta-2,4,6-trien-1-one, MANGANESE (II) ION, ... | | Authors: | Zhou, D, Wlodawer, A. | | Deposit date: | 2011-12-09 | | Release date: | 2012-03-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of the reverse transcriptase-associated ribonuclease H domain of xenotropic murine leukemia-virus related virus.
J.Struct.Biol., 177, 2012
|
|
3PGA
 
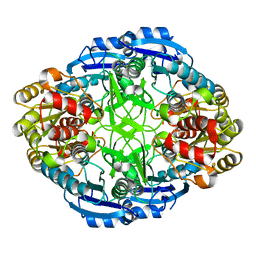 | | STRUCTURAL CHARACTERIZATION OF PSEUDOMONAS 7A GLUTAMINASE-ASPARAGINASE | | Descriptor: | GLUTAMINASE-ASPARAGINASE | | Authors: | Lubkowski, J, Wlodawer, A, Ammon, H.L, Copeland, T.D, Swain, A.L. | | Deposit date: | 1994-07-19 | | Release date: | 1994-12-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization of Pseudomonas 7A glutaminase-asparaginase.
Biochemistry, 33, 1994
|
|
5E0S
 
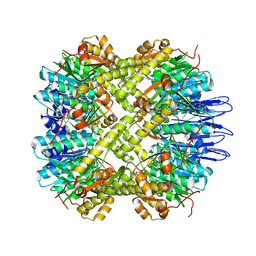 | | crystal structure of the active form of the proteolytic complex clpP1 and clpP2 | | Descriptor: | ATP-dependent Clp protease proteolytic subunit 1, ATP-dependent Clp protease proteolytic subunit 2 | | Authors: | LI, M, Wlodawer, A, Maurizi, M. | | Deposit date: | 2015-09-29 | | Release date: | 2016-02-17 | | Last modified: | 2016-04-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and Functional Properties of the Active Form of the Proteolytic Complex, ClpP1P2, from Mycobacterium tuberculosis.
J.Biol.Chem., 291, 2016
|
|
9AVQ
 
 | | Crystal structure of SARS-CoV-2 main protease A191T mutant in complex with an inhibitor Nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER | | Authors: | Bulut, H, Hattori, S, Hayashi, H, Hasegawa, K, Li, M, Wlodawer, A, Tamamura, H, Mitsuya, H. | | Deposit date: | 2024-03-04 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural and virologic mechanism of emergence of main protease inhibitor-resistance in SARS-CoV-2 as selected with main protease inhibitors
To Be Published
|
|
9ARQ
 
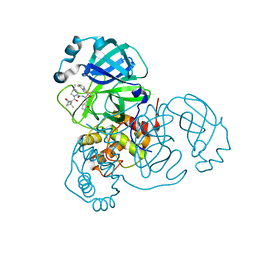 | | Crystal structure of SARS-CoV-2 main protease (authentic protein) in complex with an inhibitor TKB-245 | | Descriptor: | (1R,2S,5S)-N-{(1S,2S)-1-(4-fluoro-1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Bulut, H, Hattori, S, Hayashi, H, Hasegawa, K, Li, M, Wlodawer, A, Tamamura, H, Mitsuya, H. | | Deposit date: | 2024-02-23 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and virologic mechanism of emergence of main protease inhibitor-resistance in SARS-CoV-2 as selected with main protease inhibitors
To Be Published
|
|
9ARS
 
 | | Crystal structure of SARS-CoV-2 main protease E166V mutant in complex with an inhibitor TKB-245 | | Descriptor: | (1R,2S,5S)-N-{(1S,2S)-1-(4-fluoro-1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Bulut, H, Hattori, S, Hayashi, H, Hasegawa, K, Li, M, Wlodawer, A, Misumi, S, Tamamura, H, Mitsuya, H. | | Deposit date: | 2024-02-23 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and virologic mechanism of emergence of main protease inhibitor-resistance in SARS-CoV-2 as selected with main protease inhibitors
To Be Published
|
|
