2LO4
 
 | |
4WYQ
 
 | | Crystal structure of the Dicer-TRBP interface | | Descriptor: | Endoribonuclease Dicer, Poly(UNK), RISC-loading complex subunit TARBP2 | | Authors: | Wilson, R.C, Doudna, J.A. | | Deposit date: | 2014-11-18 | | Release date: | 2014-12-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Dicer-TRBP Complex Formation Ensures Accurate Mammalian MicroRNA Biogenesis.
Mol.Cell, 57, 2015
|
|
3F92
 
 | | Crystal structure of ubiquitin-conjugating enzyme E2-25kDa (Huntington Interacting Protein 2) M172A mutant crystallized at pH 8.5 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, CALCIUM ION, ... | | Authors: | Wilson, R.C, Hughes, R.C, Flatt, J.W, Meehan, E.J, Ng, J.D, Twigg, P.D. | | Deposit date: | 2008-11-13 | | Release date: | 2008-11-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structure of full-length ubiquitin-conjugating enzyme E2-25K (huntingtin-interacting protein 2).
Acta Crystallogr.,Sect.F, 65, 2009
|
|
2AV5
 
 | | Crystal structure of Pyrococcus furiosus Pop5, an archaeal Ribonuclease P protein | | Descriptor: | Ribonuclease P protein component 2 | | Authors: | Wilson, R.C, Bohlen, C.J, Foster, M.P, Bell, C.E. | | Deposit date: | 2005-08-29 | | Release date: | 2006-01-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structure of Pfu Pop5, an archaeal RNase P protein.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
5I4A
 
 | | X-ray crystal structure of Marinitoga piezophila Argonaute in complex with 5' OH guide RNA | | Descriptor: | Argonaute protein, RNA (5'-R(*UP*AP*UP*AP*CP*AP*AP*CP*CP*UP*AP*CP*UP*U)-3') | | Authors: | Doxzen, K.W, Kaya, E, Knoll, K.R, Wilson, R.C, Strutt, S.C, Kranzusch, P.J, Doudna, J.A. | | Deposit date: | 2016-02-11 | | Release date: | 2016-03-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | A bacterial Argonaute with noncanonical guide RNA specificity.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3BQ1
 
 | | Insertion ternary complex of Dbh DNA polymerase | | Descriptor: | 2'-3'-DIDEOXYGUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*DGP*DAP*DAP*DGP*DCP*DCP*DGP*DGP*DCP*DG)-3'), ... | | Authors: | Pata, J.D, Wilson, R.C. | | Deposit date: | 2007-12-19 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into the generation of single-base deletions by the Y family DNA polymerase dbh.
Mol.Cell, 29, 2008
|
|
3BQ0
 
 | | Pre-insertion binary complex of Dbh DNA polymerase | | Descriptor: | CALCIUM ION, DNA (5'-D(*DGP*DAP*DAP*DGP*DCP*DCP*DGP*DGP*DCP*DG)-3'), DNA (5'-D(*DT*DTP*DCP*DCP*DGP*DCP*DCP*DCP*DGP*DGP*DCP*DTP*DTP*DCP*DC)-3'), ... | | Authors: | Pata, J.D, Wilson, R.C. | | Deposit date: | 2007-12-19 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into the generation of single-base deletions by the Y family DNA polymerase dbh.
Mol.Cell, 29, 2008
|
|
3BQ2
 
 | | Post-insertion binary complex of Dbh DNA polymerase | | Descriptor: | CALCIUM ION, DNA (5'-D(*DGP*DAP*DAP*DGP*DCP*DCP*DGP*DGP*DCP*DGP*DG)-3'), DNA (5'-D(*DTP*DTP*DCP*DCP*DGP*DCP*DCP*DCP*DGP*DGP*DCP*DTP*DTP*DCP*DC)-3'), ... | | Authors: | Pata, J.D, Wilson, R.C. | | Deposit date: | 2007-12-19 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into the generation of single-base deletions by the Y family DNA polymerase dbh.
Mol.Cell, 29, 2008
|
|
3E46
 
 | | Crystal structure of ubiquitin-conjugating enzyme E2-25kDa (Huntington interacting protein 2) M172A mutant | | Descriptor: | CALCIUM ION, Ubiquitin-conjugating enzyme E2-25 kDa | | Authors: | Hughes, R.C, Wilson, R.C, Flatt, J.W, Meehan, E.J, Ng, J.D, Twigg, P.D. | | Deposit date: | 2008-08-09 | | Release date: | 2008-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure of full-length ubiquitin-conjugating enzyme E2-25K (huntingtin-interacting protein 2).
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3QZ8
 
 | | TT-4 ternary complex of Dpo4 | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*GP*GP*CP*AP*CP*TP*GP*AP*TP*CP*AP*GP*G)-3', 5'-D(*TP*TP*AP*CP*GP*CP*CP*TP*TP*GP*AP*TP*CP*AP*GP*TP*GP*CP*C)-3', ... | | Authors: | Pata, J.D, Wu, Y, Wilson, R.C. | | Deposit date: | 2011-03-04 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | The y-family DNA polymerase dpo4 uses a template slippage mechanism to create single-base deletions.
J.Bacteriol., 193, 2011
|
|
3QZ7
 
 | | T-3 ternary complex of Dpo4 | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*GP*GP*CP*AP*CP*TP*GP*AP*TP*CP*GP*GP*G)-3', 5'-D(*TP*TP*AP*CP*GP*CP*CP*TP*CP*GP*AP*TP*CP*AP*GP*TP*GP*CP*C)-3', ... | | Authors: | Pata, J.D, Wu, Y, Wilson, R.C. | | Deposit date: | 2011-03-04 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | The y-family DNA polymerase dpo4 uses a template slippage mechanism to create single-base deletions.
J.Bacteriol., 193, 2011
|
|
4NLG
 
 | | Y-family DNA polymerase chimera Dbh-Dpo4(243-245)-Dbh | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*GP*GP*CP*AP*CP*TP*GP*AP*TP*CP*GP*GP*G)-3', 5'-D(*TP*TP*AP*CP*GP*CP*CP*CP*TP*GP*AP*TP*CP*AP*GP*TP*GP*CP*C)-3', ... | | Authors: | Mukherjee, P, Wilson, R.C, Pata, J.D. | | Deposit date: | 2013-11-14 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Three Residues of the Interdomain Linker Determine the Conformation and Single-base Deletion Fidelity of Y-family Translesion Polymerases.
J.Biol.Chem., 289, 2014
|
|
1YQV
 
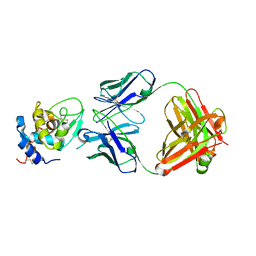 | | The crystal structure of the antibody Fab HyHEL5 complex with lysozyme at 1.7A resolution | | Descriptor: | Hen Egg White Lysozyme, HyHEL-5 Antibody Heavy Chain, HyHEL-5 Antibody Light Chain | | Authors: | Cohen, G.H, Silverton, E.W, Padlan, E.A, Dyda, F, Wibbenmeyer, J.A, Wilson, R.C, Davies, D.R. | | Deposit date: | 2005-02-02 | | Release date: | 2005-04-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Water molecules in the antibody-antigen interface of the structure of the Fab HyHEL-5-lysozyme complex at 1.7 A resolution: comparison with results from isothermal titration calorimetry.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
4F4W
 
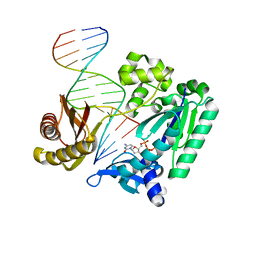 | | Y-family DNA polymerase chimera Dbh-Dpo4-Dpo4 #1 | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*GP*GP*CP*AP*CP*TP*GP*AP*TP*CP*GP*GP*C)-3'), ... | | Authors: | Pata, J.D, Wilson, R.C. | | Deposit date: | 2012-05-11 | | Release date: | 2013-01-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Y-family polymerase conformation is a major determinant of fidelity and translesion specificity.
Structure, 21, 2013
|
|
4F4X
 
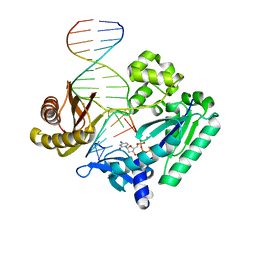 | | Y-family DNA polymerase chimera Dbh-Dpo4-Dpo4 #2 | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*GP*GP*CP*AP*CP*TP*GP*AP*TP*CP*GP*GP*GP*C)-3'), ... | | Authors: | Pata, J.D, Wilson, R.C. | | Deposit date: | 2012-05-11 | | Release date: | 2013-01-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | Y-family polymerase conformation is a major determinant of fidelity and translesion specificity.
Structure, 21, 2013
|
|
4F4Y
 
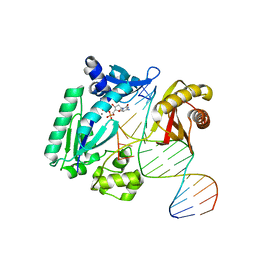 | | Y-family DNA polymerase chimera Dbh-Dpo4-Dbh | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*GP*GP*CP*AP*CP*TP*GP*AP*TP*CP*GP*GP*GP*CP*CP*C)-3'), ... | | Authors: | Pata, J.D, Wilson, R.C. | | Deposit date: | 2012-05-11 | | Release date: | 2013-01-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.338 Å) | | Cite: | Y-family polymerase conformation is a major determinant of fidelity and translesion specificity.
Structure, 21, 2013
|
|
4F4Z
 
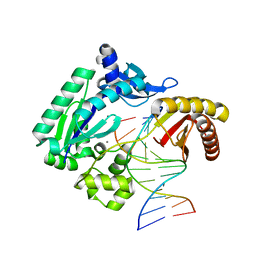 | | Y-family DNA polymerase chimera Dpo4-Dpo4-Dbh | | Descriptor: | CALCIUM ION, DNA (5'-D(*AP*GP*GP*GP*GP*GP*AP*AP*GP*CP*CP*G)-3'), DNA (5'-D(*TP*TP*CP*CP*GP*CP*CP*CP*GP*GP*CP*TP*TP*CP*CP*CP*CP*CP*T)-3'), ... | | Authors: | Pata, J.D, Wilson, R.C. | | Deposit date: | 2012-05-11 | | Release date: | 2013-01-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.305 Å) | | Cite: | Y-family polymerase conformation is a major determinant of fidelity and translesion specificity.
Structure, 21, 2013
|
|
4F50
 
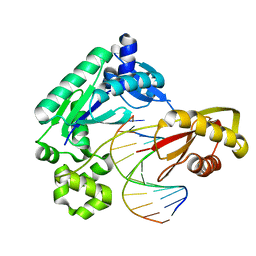 | | Y-family DNA polymerase chimera Dbh-Dbh-Dpo4 | | Descriptor: | CALCIUM ION, DNA (5'-D(*CP*CP*GP*CP*CP*T*GP*GP*CP*TP*TP*CP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*AP*AP*GP*CP*CP*GP*GP*C)-3'), ... | | Authors: | Pata, J.D, Wilson, R.C. | | Deposit date: | 2012-05-11 | | Release date: | 2013-01-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.215 Å) | | Cite: | Y-family polymerase conformation is a major determinant of fidelity and translesion specificity.
Structure, 21, 2013
|
|
4HYK
 
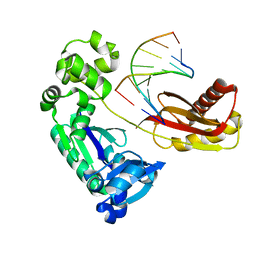 | | Dbh Ternary Complex (substrates partially disordered) | | Descriptor: | DNA (5'-D(*GP*GP*GP*AP*AP*GP*CP*CP*GP*G)-3'), DNA (5'-D(*TP*TP*CP*CP*GP*CP*CP*CP*CP*GP*GP*CP*TP*TP*CP*CP*C)-3'), DNA polymerase IV | | Authors: | Pata, J.D, Wilson, R.C. | | Deposit date: | 2012-11-13 | | Release date: | 2013-01-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Y-family polymerase conformation is a major determinant of fidelity and translesion specificity.
Structure, 21, 2013
|
|
