5OD5
 
 | | Periplasmic binding protein CeuE complexed with a synthetic catalyst | | Descriptor: | 2,5,8,11,14,17,20,23-OCTAOXAPENTACOSAN-25-OL, 4-(aminomethyl)-~{N}-(pyridin-2-ylmethyl)benzenesulfonamide, Azotochelin, ... | | Authors: | Duhme-Klair, A.K, Raines, D.J, Clarke, J.E, Blagova, E.V, Dodson, E.J, Wilson, K.S. | | Deposit date: | 2017-07-04 | | Release date: | 2018-08-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Redox-switchable siderophore anchor enables reversible artificial metalloenzyme assembly
Nat Catal, 2018
|
|
4CFY
 
 | | SAVINASE CRYSTAL STRUCTURES FOR COMBINED SINGLE CRYSTAL DIFFRACTION AND POWDER DIFFRACTION ANALYSIS | | Descriptor: | CALCIUM ION, SODIUM ION, SUBTILISIN SAVINASE | | Authors: | Frankaer, C.G, Moroz, O.V, Turkenburg, J.P, Aspmo, S.I, Thymark, M, Friis, E.P, Stahla, K, Nielsen, J.E, Wilson, K.S, Harris, P. | | Deposit date: | 2013-11-19 | | Release date: | 2014-04-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Analysis of an Industrial Production Suspension of Bacillus Lentus Subtilisin Crystals by Powder Diffraction: A Powerful Quality-Control Tool.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4CFZ
 
 | | SAVINASE CRYSTAL STRUCTURES FOR COMBINED SINGLE CRYSTAL DIFFRACTION AND POWDER DIFFRACTION ANALYSIS | | Descriptor: | CALCIUM ION, SODIUM ION, SUBTILISIN SAVINASE, ... | | Authors: | Frankaer, C.G, Moroz, O.V, Turkenburg, J.P, Aspmo, S.I, Thymark, M, Friis, E.P, Stahla, K, Nielsen, J.E, Wilson, K.S, Harris, P. | | Deposit date: | 2013-11-19 | | Release date: | 2014-04-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Analysis of an Industrial Production Suspension of Bacillus Lentus Subtilisin Crystals by Powder Diffraction: A Powerful Quality-Control Tool.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1XSO
 
 | | THREE-DIMENSIONAL STRUCTURE OF XENOPUS LAEVIS CU,ZN SUPEROXIDE DISMUTASE B DETERMINED BY X-RAY CRYSTALLOGRAPHY AT 1.5 ANGSTROMS RESOLUTION | | Descriptor: | COPPER (II) ION, COPPER,ZINC SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Djinovic Carugo, K, Coda, A, Battistoni, A, Carri, M.T, Polticelli, F, Desideri, A, Rotilio, G, Wilson, K.S, Bolognesi, M. | | Deposit date: | 1995-03-14 | | Release date: | 1995-07-10 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Three-dimensional structure of Xenopus laevis Cu,Zn superoxide dismutase b determined by X-ray crystallography at 1.5 A resolution.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
4DK2
 
 | |
4DL8
 
 | | Crystal structure of Trypanosoma brucei dUTPase with dUMP, planar [AlF3-OPO3] transition state analogue, Mg2+, and Na+ | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, ALUMINUM FLUORIDE, Deoxyuridine triphosphatase, ... | | Authors: | Hemsworth, G.R, Gonzalez-Pacanowska, D, Wilson, K.S. | | Deposit date: | 2012-02-06 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | On the catalytic mechanism of dimeric dUTPases.
Biochem.J., 456, 2013
|
|
4DK4
 
 | | Crystal Structure of Trypanosoma brucei dUTPase with dUpNp, Ca2+ and Na+ | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-DIPHOSPHATE, CALCIUM ION, Deoxyuridine triphosphatase, ... | | Authors: | Hemsworth, G.R, Gonzalez-Pacanowska, D, Wilson, K.S. | | Deposit date: | 2012-02-03 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | On the catalytic mechanism of dimeric dUTPases.
Biochem.J., 456, 2013
|
|
6F1J
 
 | | Structure of a Talaromyces pinophilus GH62 Arabinofuranosidase in complex with AraDNJ at 1.25A resolution | | Descriptor: | 1,4-DIDEOXY-1,4-IMINO-L-ARABINITOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Moroz, O.V, Sobala, L, Blagova, E, Coyle, T, Morkeberg Krogh, K.B.R, Wei, P, Stubbs, K, Wilson, K.S, Davies, G.J. | | Deposit date: | 2017-11-22 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structure of a Talaromyces pinophilus GH62 arabinofuranosidase in complex with AraDNJ at 1.25 angstrom resolution.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6FHV
 
 | | Crystal structure of Penicillium oxalicum Glucoamylase | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Roth, C, Moroz, O.V, Ariza, A, Friis, E.P, Davies, G.J, Wilson, K.S. | | Deposit date: | 2018-01-15 | | Release date: | 2018-05-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insight into industrially relevant glucoamylases: flexible positions of starch-binding domains.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6FHN
 
 | | Structural dynamics and catalytic properties of a multi-modular xanthanase (Pt derivative) | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Moroz, O.V, Jensen, P.F, McDonald, S.P, McGregor, N, Blagova, E, Comamala, G, Segura, D.R, Anderson, L, Vasu, S.M, Rao, V.P, Giger, L, Monrad, R.N, Svendsen, A, Nielsen, J.E, Henrissat, B, Davies, G.J, Brumer, H, Rand, K, Wilson, K.S. | | Deposit date: | 2018-01-15 | | Release date: | 2018-08-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Dynamics and Catalytic Properties of a Multimodular Xanthanase
Acs Catalysis, 2018
|
|
6FHW
 
 | | Structure of Hormoconis resinae Glucoamylase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Roth, C, Moroz, O.V, Ariza, A, Friis, E.P, Davies, G.J, Wilson, K.S. | | Deposit date: | 2018-01-15 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural insight into industrially relevant glucoamylases: flexible positions of starch-binding domains.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6FHJ
 
 | | Structural dynamics and catalytic properties of a multi-modular xanthanase, native. | | Descriptor: | CALCIUM ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Moroz, O.V, Jensen, P.F, McDonald, S.P, McGregor, N, Blagova, E, Comamala, G, Segura, D.R, Anderson, L, Vasu, S.M, Rao, V.P, Giger, L, Monrad, R.N, Svendsen, A, Nielsen, J.E, Henrissat, B, Davies, G.J, Brumer, H, Rand, K, Wilson, K.S. | | Deposit date: | 2018-01-14 | | Release date: | 2018-08-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural Dynamics and Catalytic Properties of a Multimodular Xanthanase
Acs Catalysis, 2018
|
|
6GXV
 
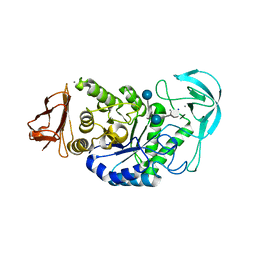 | | Amylase in complex with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, A-amylase, CALCIUM ION, ... | | Authors: | Agirre, J, Moroz, O, Meier, S, Brask, J, Munch, A, Hoff, T, Andersen, C, Wilson, K.S, Davies, G.J. | | Deposit date: | 2018-06-27 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | The structure of the AliC GH13 alpha-amylase from Alicyclobacillus sp. reveals the accommodation of starch branching points in the alpha-amylase family.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6GYA
 
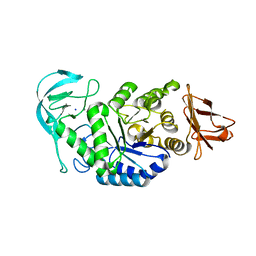 | | Amylase in complex with branched ligand | | Descriptor: | A-amylase, CALCIUM ION, SODIUM ION, ... | | Authors: | Agirre, J, Moroz, O, Meier, S, Brask, J, Munch, A, Hoff, T, Andersen, C, Wilson, K.S, Davies, G.J. | | Deposit date: | 2018-06-28 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The structure of the AliC GH13 alpha-amylase from Alicyclobacillus sp. reveals the accommodation of starch branching points in the alpha-amylase family.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6FRV
 
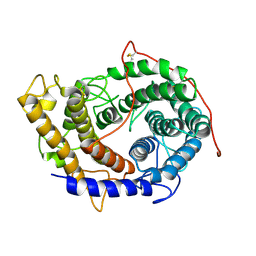 | | Structure of the catalytic domain of Aspergillus niger Glucoamylase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glucoamylase, ... | | Authors: | Roth, C, Moroz, O.V, Ariza, A, Friis, E.P, Davies, G.J, Wilson, K.S. | | Deposit date: | 2018-02-16 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insight into industrially relevant glucoamylases: flexible positions of starch-binding domains.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6G21
 
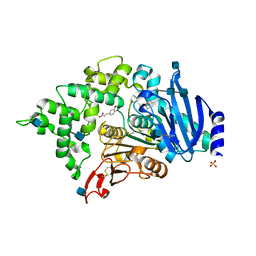 | | Crystal structure of an esterase from Aspergillus oryzae | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, ... | | Authors: | Moroz, O.V, Blagova, E, Davies, G.J, Wilson, K.S. | | Deposit date: | 2018-03-22 | | Release date: | 2018-05-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of an esterase from Aspergillus oryzae
To Be Published
|
|
1S3T
 
 | | BORATE INHIBITED BACILLUS PASTEURII UREASE CRYSTAL STRUCTURE | | Descriptor: | BORIC ACID, NICKEL (II) ION, SULFATE ION, ... | | Authors: | Benini, S, Rypniewski, W.R, Wilson, K.S, Ciurli, S, Mangani, S. | | Deposit date: | 2004-01-14 | | Release date: | 2004-04-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular Details of Urease Inhibition by Boric Acid: Insights into the Catalytic Mechanism.
J.Am.Chem.Soc., 126, 2004
|
|
2NAD
 
 | | HIGH RESOLUTION STRUCTURES OF HOLO AND APO FORMATE DEHYDROGENASE | | Descriptor: | AZIDE ION, NAD-DEPENDENT FORMATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Lamzin, V.S, Dauter, Z, Popov, V.O, Harutyunyan, E.H, Wilson, K.S. | | Deposit date: | 1994-07-06 | | Release date: | 1995-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | High resolution structures of holo and apo formate dehydrogenase.
J.Mol.Biol., 236, 1994
|
|
2NAC
 
 | | HIGH RESOLUTION STRUCTURES OF HOLO AND APO FORMATE DEHYDROGENASE | | Descriptor: | NAD-DEPENDENT FORMATE DEHYDROGENASE, SULFATE ION | | Authors: | Lamzin, V.S, Dauter, Z, Popov, V.O, Harutyunyan, E.H, Wilson, K.S. | | Deposit date: | 1994-07-06 | | Release date: | 1995-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High resolution structures of holo and apo formate dehydrogenase.
J.Mol.Biol., 236, 1994
|
|
2YJQ
 
 | | Structure of a Paenibacillus Polymyxa Xyloglucanase from Glycoside Hydrolase Family 44 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CEL44C, ... | | Authors: | Ariza, A, Eklof, J.M, Spadiut, O, Offen, W.A, Roberts, S.M, Besenmatter, W, Friis, E.P, Skjot, M, Wilson, K.S, Brumer, H, Davies, G. | | Deposit date: | 2011-05-23 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure and Activity of Paenibacillus Polymyxa Xyloglucanase from Glycoside Hydrolase Family 44.
J.Biol.Chem., 286, 2011
|
|
2Y1T
 
 | | Bacillus subtilis prophage dUTPase in complex with dUDP | | Descriptor: | DEOXYURIDINE-5'-DIPHOSPHATE, SPBC2 PROPHAGE-DERIVED DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE YOSS | | Authors: | Garcia-Nafria, J, Harkiolaki, M, Persson, R, Fogg, M.J, Wilson, K.S. | | Deposit date: | 2010-12-10 | | Release date: | 2011-02-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The Structure of Bacillus Subtilis Sp Beta Prophage Dutpase and its Complexes with Two Nucleotides
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2Y8C
 
 | | Plasmodium falciparum dUTPase in complex with a trityl ligand | | Descriptor: | (2S)-2-[(2,4-dioxopyrimidin-1-yl)methyl]-N-(2-hydroxyethyl)-4-trityloxy-butanamide, DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE, SULFATE ION | | Authors: | Baragana, B, McCarthy, O, Sanchez, P, Bosch, C, Kaiser, M, Brun, R, Whittingham, J.L, Roberts, S, Zhou, X.-X, Wilson, K.S, Johansson, N.G, Gonzalez-Pacanowska, D, Gilbert, I.H. | | Deposit date: | 2011-02-04 | | Release date: | 2012-03-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Beta-Branched Acyclic Nucleoside Analogues as Inhibitors of Plasmodium Falciparum Dutpase
Bioorg.Med.Chem., 19, 2011
|
|
2YB0
 
 | | The Crystal Structure of Leishmania major dUTPase in complex deoxyuridine | | Descriptor: | 2'-DEOXYURIDINE, DUTPASE, SULFATE ION | | Authors: | Hemsworth, G.R, Moroz, O.V, Fogg, M.J, Scott, B, Bosch-Navarrete, C, Gonzalez-Pacanowska, D, Wilson, K.S. | | Deposit date: | 2011-02-25 | | Release date: | 2011-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | The Crystal Structure of the Leishmania Major Deoxyuridine Triphosphate Nucleotidohydrolase in Complex with Nucleotide Analogues, Dump, and Deoxyuridine.
J.Biol.Chem., 286, 2011
|
|
2YAZ
 
 | | The Crystal Structure of Leishmania major dUTPase in complex dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, DUTPASE, MAGNESIUM ION, ... | | Authors: | Hemsworth, G.R, Moroz, O.V, Fogg, M.J, Scott, B, Bosch-Navarrete, C, Gonzalez-Pacanowska, D, Wilson, K.S. | | Deposit date: | 2011-02-25 | | Release date: | 2011-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Crystal Structure of the Leishmania Major Deoxyuridine Triphosphate Nucleotidohydrolase in Complex with Nucleotide Analogues, Dump, and Deoxyuridine.
J.Biol.Chem., 286, 2011
|
|
2YOH
 
 | | Plasmodium falciparum thymidylate kinase in complex with a urea-alpha- deoxythymidine inhibitor | | Descriptor: | 1-[[(2R,3S,5S)-5-[5-methyl-2,4-bis(oxidanylidene)pyrimidin-1-yl]-3-oxidanyl-oxolan-2-yl]methyl]-3-(4-nitrophenyl)urea, THYMIDYLATE KINASE | | Authors: | Huaqing, C, Carrero-Lerida, J, Silva, A.P.G, Whittingham, J.L, Brannigan, J.A, Ruiz-Perez, L.M, Read, K.D, Wilson, K.S, Gonzalez-Pacanowska, D, Gilbert, I.H. | | Deposit date: | 2012-10-24 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Synthesis and Evaluation of Alpha-Thymidine Analogues as Novel Antimalarials.
J.Med.Chem., 55, 2012
|
|
