8BF6
 
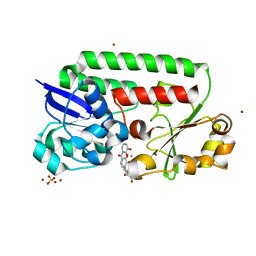 | | X-ray structure of the CeuE Homologue from Parageobacillus thermoglucosidasius - azotochelin complex | | Descriptor: | ABC transporter, Azotochelin, FE (III) ION, ... | | Authors: | Wilson, K.S, Duhme-Klair, A.-K, Blagova, E.V, Miller, A, Booth, R, Dodson, E.J. | | Deposit date: | 2022-10-24 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.969 Å) | | Cite: | Thermostable homologues of the periplasmic siderophore-binding protein CeuE from Geobacillus stearothermophilus and Parageobacillus thermoglucosidasius.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8B7X
 
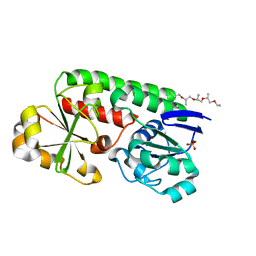 | | X-ray structure of the CeuE Homologue from Geobacillus stearothermophilus - apo form. | | Descriptor: | O-(O-(2-AMINOPROPYL)-O'-(2-METHOXYETHYL)POLYPROPYLENE GLYCOL 500), SULFATE ION, Siderophore ABC transporter substrate-binding protein | | Authors: | Wilson, K.S, Duhme-Klair, A.K, Blagova, E.V, Bennett, M. | | Deposit date: | 2022-10-03 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Thermostable homologues of the periplasmic siderophore-binding protein CeuE from Geobacillus stearothermophilus and Parageobacillus thermoglucosidasius.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
3ZHC
 
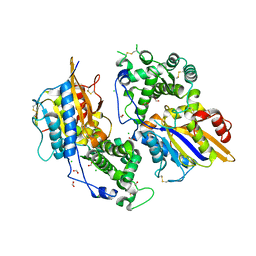 | | Structure of the phytase from Citrobacter braakii at 2.3 angstrom resolution. | | Descriptor: | CHLORIDE ION, FORMIC ACID, PHYTASE | | Authors: | Wilson, K.S, Ariza, A, Sanchez-Romero, I, Skjot, M, Vind, J, DeMaria, L, Skov, L.K, Sanchez-Ruiz, J.M. | | Deposit date: | 2012-12-20 | | Release date: | 2013-08-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanism of Protein Kinetic Stabilization by Engineered Disulfide Crosslinks
Plos One, 8, 2013
|
|
4RVE
 
 | | THE CRYSTAL STRUCTURE OF ECORV ENDONUCLEASE AND OF ITS COMPLEXES WITH COGNATE AND NON-COGNATE DNA SEGMENTS | | Descriptor: | DNA (5'-D(*GP*GP*GP*AP*TP*AP*TP*CP*CP*C)-3'), PROTEIN (ECO RV (E.C.3.1.21.4)) | | Authors: | Winkler, F.K, Banner, D.W, Oefner, C, Tsernoglou, D, Brown, R.S, Heathman, S.P, Bryan, R.K, Martin, P.D, Petratos, K, Wilso, K.S. | | Deposit date: | 1993-02-18 | | Release date: | 1993-04-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of EcoRV endonuclease and of its complexes with cognate and non-cognate DNA fragments.
EMBO J., 12, 1993
|
|
2D30
 
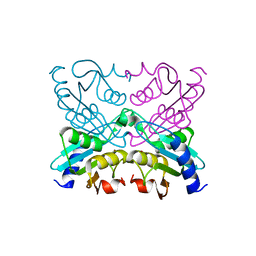 | | Crystal Structure of Cytidine Deaminase Cdd-2 (BA4525) from Bacillus Anthracis at 2.40A Resolution | | Descriptor: | ZINC ION, cytidine deaminase | | Authors: | Levdikov, V.M, Blagova, E.V, Fogg, M.J, Brannigan, J.A, Moroz, O.V, Wilkinson, A.J, Wilson, K.S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-09-21 | | Release date: | 2006-09-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Cytidine Deaminase Cdd-2 (BA4525) from Bacillus Anthracis at 2.40A Resolution
To be Published
|
|
4UZU
 
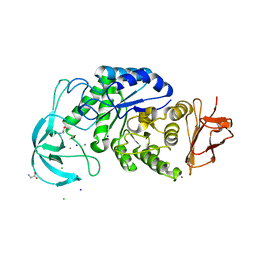 | | Three-dimensional structure of a variant `Termamyl-like' Geobacillus stearothermophilus alpha-amylase at 1.9 A resolution | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Offen, W.A, Anderson, C, Borchert, T.V, Wilson, K.S, Davies, G.J. | | Deposit date: | 2014-09-09 | | Release date: | 2015-01-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Three-Dimensional Structure of a Variant `Termamyl-Like' Geobacillus Stearothermophilus Alpha-Amylase at 1.9 A Resolution
Acta Crystallogr.,Sect.F, 71, 2015
|
|
1UBP
 
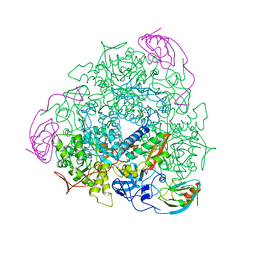 | | CRYSTAL STRUCTURE OF UREASE FROM BACILLUS PASTEURII INHIBITED WITH BETA-MERCAPTOETHANOL AT 1.65 ANGSTROMS RESOLUTION | | Descriptor: | BETA-MERCAPTOETHANOL, NICKEL (II) ION, UREASE | | Authors: | Benini, S, Rypniewski, W.R, Wilson, K.S, Ciurli, S, Mangani, S. | | Deposit date: | 1998-01-21 | | Release date: | 1999-03-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The complex of Bacillus pasteurii urease with beta-mercaptoethanol from X-ray data at 1.65-A resolution
J.Biol.Inorg.Chem., 3, 1998
|
|
4V20
 
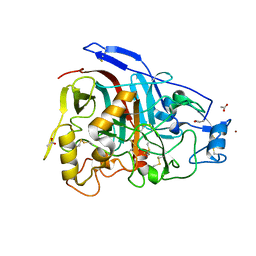 | | The 3-D structure of the cellobiohydrolase, Cel7A, from Aspergillus fumigatus, disaccharide complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CELLOBIOHYDROLASE, ... | | Authors: | Moroz, O.V, Maranta, M, Shaghasi, T, Harris, P.V, Wilson, K.S, Davies, G.J. | | Deposit date: | 2014-10-05 | | Release date: | 2015-01-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Three-Dimensional Structure of the Cellobiohydrolase Cel7A from Aspergillus Fumigatus at 1.5 A Resolution
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4V6V
 
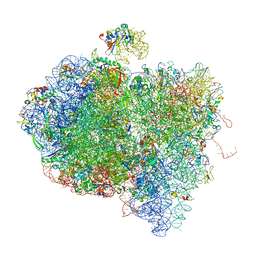 | | Tetracycline resistance protein Tet(O) bound to the ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Li, W, Atkinson, G.C, Thakor, N.S, Allas, U, Lu, C, Chan, K.Y, Tenson, T, Schulten, K, Wilson, K.S, Hauryliuk, V, Frank, J. | | Deposit date: | 2013-02-25 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (9.8 Å) | | Cite: | Mechanism of tetracycline resistance by ribosomal protection protein Tet(O).
Nat Commun, 4, 2013
|
|
2WC8
 
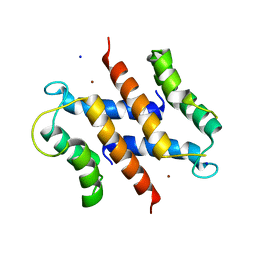 | | S100A12 complex with zinc in the absence of calcium | | Descriptor: | CITRIC ACID, PROTEIN S100-A12, SODIUM ION, ... | | Authors: | Moroz, O.V, Blagova, E.V, Wilkinson, A.J, Wilson, K.S, Bronstein, I.B. | | Deposit date: | 2009-03-10 | | Release date: | 2009-06-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The Crystal Structures of Human S100A12 in Apo Form and in Complex with Zinc: New Insights Into S100A12 Oligomerisation.
J.Mol.Biol., 391, 2009
|
|
6QPP
 
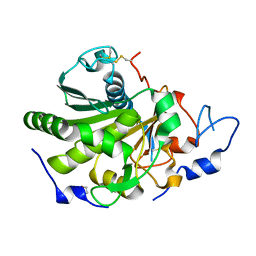 | | Rhizomucor miehei lipase propeptide complex, native | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Lipase | | Authors: | Moroz, O.V, Blagova, E, Reiser, V, Saikia, R, Dalal, S, Jorgensen, C.I, Baunsgaard, L, Andersen, B, Svendsen, A, Wilson, K.S. | | Deposit date: | 2019-02-14 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Novel Inhibitory Function of theRhizomucor mieheiLipase Propeptide and Three-Dimensional Structures of Its Complexes with the Enzyme.
Acs Omega, 4, 2019
|
|
6QPR
 
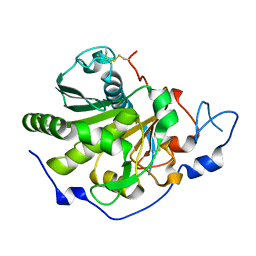 | | Rhizomucor miehei lipase propeptide complex, Ser95/Ile96 deletion mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lipase | | Authors: | Moroz, O.V, Blagova, E, Reiser, V, Saikia, R, Dalal, S, Jorgensen, C.I, Baunsgaard, L, Andersen, B, Svendsen, A, Wilson, K.S. | | Deposit date: | 2019-02-14 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Novel Inhibitory Function of theRhizomucor mieheiLipase Propeptide and Three-Dimensional Structures of Its Complexes with the Enzyme.
Acs Omega, 4, 2019
|
|
7NSN
 
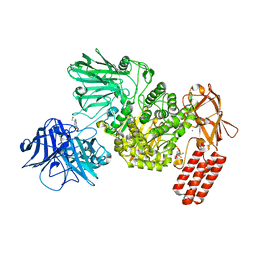 | | Multi-domain GH92 alpha-1,2-mannosidase from Neobacillus novalis: mannoimidazole complex | | Descriptor: | (5R,6R,7S,8R)-5-(HYDROXYMETHYL)-5,6,7,8-TETRAHYDROIMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Kolaczkowski, B.M, Moroz, O.V, Blagova, E, Davies, G.J, Wilson, K.S, Moeler, M.S, Meyer, A.S, Westh, P, Jensen, K, Krogh, K.B.R.M. | | Deposit date: | 2021-03-08 | | Release date: | 2022-09-21 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural and functional characterization of a multi-domain GH92 alpha-1,2-mannosidase from Neobacillus novalis.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
4DKB
 
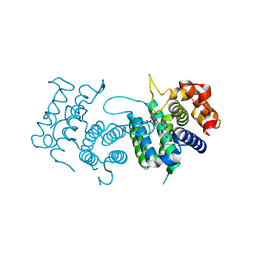 | | Crystal Structure of Trypanosoma brucei dUTPase with dUpNp and Ca2+ | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-DIPHOSPHATE, CALCIUM ION, Deoxyuridine triphosphatase | | Authors: | Hemsworth, G.R, Gonzalez-Pacanowska, D, Wilson, K.S. | | Deposit date: | 2012-02-03 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.831 Å) | | Cite: | On the catalytic mechanism of dimeric dUTPases.
Biochem.J., 456, 2013
|
|
1KF2
 
 | | Atomic Resolution Structure of RNase A at pH 5.2 | | Descriptor: | SULFATE ION, pancreatic ribonuclease | | Authors: | Berisio, R, Sica, F, Lamzin, V.S, Wilson, K.S, Zagari, A, Mazzarella, L. | | Deposit date: | 2001-11-19 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic resolution structures of ribonuclease A at six pH values.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1KF4
 
 | | Atomic Resolution Structure of RNase A at pH 6.3 | | Descriptor: | SULFATE ION, pancreatic ribonuclease | | Authors: | Berisio, R, Sica, F, Lamzin, V.S, Wilson, K.S, Zagari, A, Mazzarella, L. | | Deposit date: | 2001-11-19 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic resolution structures of ribonuclease A at six pH values.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
4DK2
 
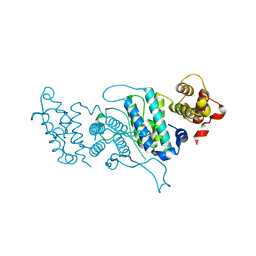 | |
1KF7
 
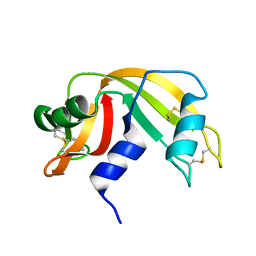 | | Atomic Resolution Structure of RNase A at pH 8.0 | | Descriptor: | pancreatic ribonuclease | | Authors: | Berisio, R, Sica, F, Lamzin, V.S, Wilson, K.S, Zagari, A, Mazzarella, L. | | Deposit date: | 2001-11-19 | | Release date: | 2001-12-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Atomic resolution structures of ribonuclease A at six pH values.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
4DL8
 
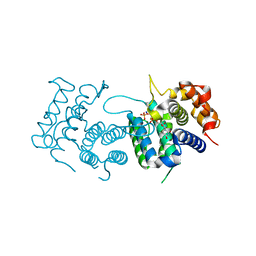 | | Crystal structure of Trypanosoma brucei dUTPase with dUMP, planar [AlF3-OPO3] transition state analogue, Mg2+, and Na+ | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, ALUMINUM FLUORIDE, Deoxyuridine triphosphatase, ... | | Authors: | Hemsworth, G.R, Gonzalez-Pacanowska, D, Wilson, K.S. | | Deposit date: | 2012-02-06 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | On the catalytic mechanism of dimeric dUTPases.
Biochem.J., 456, 2013
|
|
4DK4
 
 | | Crystal Structure of Trypanosoma brucei dUTPase with dUpNp, Ca2+ and Na+ | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-DIPHOSPHATE, CALCIUM ION, Deoxyuridine triphosphatase, ... | | Authors: | Hemsworth, G.R, Gonzalez-Pacanowska, D, Wilson, K.S. | | Deposit date: | 2012-02-03 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | On the catalytic mechanism of dimeric dUTPases.
Biochem.J., 456, 2013
|
|
1KF8
 
 | | Atomic resolution structure of RNase A at pH 8.8 | | Descriptor: | pancreatic ribonuclease | | Authors: | Berisio, R, Sica, F, Lamzin, V.S, Wilson, K.S, Zagari, A, Mazzarella, L. | | Deposit date: | 2001-11-19 | | Release date: | 2001-12-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Atomic resolution structures of ribonuclease A at six pH values.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
2UBP
 
 | | STRUCTURE OF NATIVE UREASE FROM BACILLUS PASTEURII | | Descriptor: | NICKEL (II) ION, PROTEIN (UREASE ALPHA SUBUNIT), PROTEIN (UREASE BETA SUBUNIT), ... | | Authors: | Benini, S, Rypniewski, W.R, Wilson, K.S, Ciurli, S, Mangani, S. | | Deposit date: | 1998-11-04 | | Release date: | 1999-11-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A new proposal for urease mechanism based on the crystal structures of the native and inhibited enzyme from Bacillus pasteurii: why urea hydrolysis costs two nickels.
Structure Fold.Des., 7, 1999
|
|
6F1J
 
 | | Structure of a Talaromyces pinophilus GH62 Arabinofuranosidase in complex with AraDNJ at 1.25A resolution | | Descriptor: | 1,4-DIDEOXY-1,4-IMINO-L-ARABINITOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Moroz, O.V, Sobala, L, Blagova, E, Coyle, T, Morkeberg Krogh, K.B.R, Wei, P, Stubbs, K, Wilson, K.S, Davies, G.J. | | Deposit date: | 2017-11-22 | | Release date: | 2018-08-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structure of a Talaromyces pinophilus GH62 arabinofuranosidase in complex with AraDNJ at 1.25 angstrom resolution.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
2W6K
 
 | | The crystal structure at 1.7 A resolution of CobE, a protein from the cobalamin (vitamin B12) biosynthetic pathway | | Descriptor: | COBE, GLYCEROL, SULFATE ION | | Authors: | Vevodova, J, Smith, D, McGoldrick, H, Deery, E, Murzin, A.G, Warren, M.J, Wilson, K.S. | | Deposit date: | 2008-12-18 | | Release date: | 2008-12-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure at 1.7 A Resolution of Cobe, a Protein from the Cobalamin (Vitamin B12) Biosynthetic Pathway
To be Published
|
|
2CBF
 
 | | THE X-RAY STRUCTURE OF A COBALAMIN BIOSYNTHETIC ENZYME, COBALT PRECORRIN-4 METHYLTRANSFERASE, CBIF, FROM BACILLUS MEGATERIUM, WITH THE HIS-TAG CLEAVED OFF | | Descriptor: | COBALT-PRECORRIN-4 TRANSMETHYLASE, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Schubert, H.L, Raux, E, Woodcock, S.C, Warren, M.J, Wilson, K.S. | | Deposit date: | 1998-05-01 | | Release date: | 1999-05-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The X-ray structure of a cobalamin biosynthetic enzyme, cobalt-precorrin-4 methyltransferase.
Nat.Struct.Biol., 5, 1998
|
|
