1V0L
 
 | | Xylanase Xyn10A from Streptomyces lividans in complex with xylobio-isofagomine at pH 5.8 | | Descriptor: | ENDO-1,4-BETA-XYLANASE A, PIPERIDINE-3,4-DIOL, beta-D-xylopyranose | | Authors: | Gloster, T.M, Williams, S.J, Roberts, S, Tarling, C.A, Wicki, J, Withers, S.G, Davies, G.J. | | Deposit date: | 2004-03-31 | | Release date: | 2004-08-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Atomic Resolution Analyses of the Binding of Xylobiose-Derived Deoxynojirimycin and Isofagomine to Xylanase Xyn10A
Chem.Commun.(Camb.), 16, 2004
|
|
6F90
 
 | | Structure of the family GH92 alpha-mannosidase BT3130 from Bacteroides thetaiotaomicron in complex with Mannoimidazole (ManI) | | Descriptor: | (5R,6R,7S,8R)-5-(HYDROXYMETHYL)-5,6,7,8-TETRAHYDROIMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, Alpha-1,2-mannosidase, putative, ... | | Authors: | Thompson, A.J, Spears, R.J, Zhu, Y, Suits, M.D.L, Williams, S.J, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2017-12-13 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Bacteroides thetaiotaomicron generates diverse alpha-mannosidase activities through subtle evolution of a distal substrate-binding motif.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6FAM
 
 | | Structure of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with mannose-alpha-1,3-2-aminodeoxymannojirimycin | | Descriptor: | ACETATE ION, Glycosyl hydrolase family 71, alpha-D-mannopyranose, ... | | Authors: | Fernandes, P.Z, Petricevic, M, Sobala, L.F, Davies, G.J, Williams, S.J. | | Deposit date: | 2017-12-15 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Exploration of Strategies for Mechanism-Based Inhibitor Design for Family GH99 endo-alpha-1,2-Mannanases.
Chemistry, 24, 2018
|
|
8C54
 
 | | Cryo-EM structure of NADH bound SLA dehydrogenase RlGabD from Rhizobium leguminosarum bv. trifolii SRD1565 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Succinate semialdehyde dehydrogenase | | Authors: | Sharma, M, Meek, R.W, Armstrong, Z, Blaza, J.N, Alhifthi, A, Li, J, Goddard-Borger, E.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2023-01-06 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Molecular basis of sulfolactate synthesis by sulfolactaldehyde dehydrogenase from Rhizobium leguminosarum.
Chem Sci, 14, 2023
|
|
6F91
 
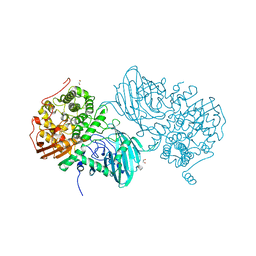 | | Structure of the family GH92 alpha-mannosidase BT3965 from Bacteroides thetaiotaomicron | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Thompson, A.J, Spears, R.J, Zhu, Y, Suits, M.D.L, Williams, S.J, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2017-12-13 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bacteroides thetaiotaomicron generates diverse alpha-mannosidase activities through subtle evolution of a distal substrate-binding motif.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
3ZIQ
 
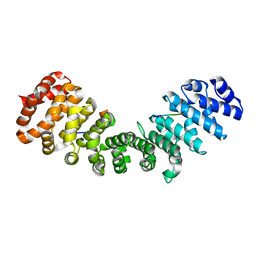 | | minor-site specific NLS (B6) | | Descriptor: | B6NLS, IMPORTIN SUBUNIT ALPHA-2 | | Authors: | Chang, C.-W, Counago, R.M, Williams, S.J, Kobe, B. | | Deposit date: | 2013-01-10 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Distinctive Conformation of Minor Site-Specific Nuclear Localization Signals Bound to Importin-Alpha
Traffic, 14, 2013
|
|
3ZIP
 
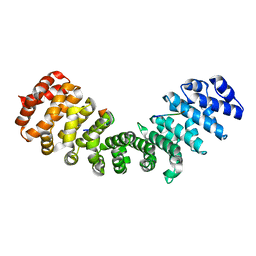 | | minor-site specific NLS (A58) | | Descriptor: | A58NLS, IMPORTIN SUBUNIT ALPHA-2 | | Authors: | Chang, C.-W, Counago, R.M, Williams, S.J, Kobe, B. | | Deposit date: | 2013-01-10 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Distinctive Conformation of Minor Site-Specific Nuclear Localization Signals Bound to Importin-Alpha
Traffic, 14, 2013
|
|
3ZIO
 
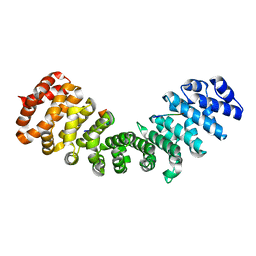 | | minor-site specific NLS (A28) | | Descriptor: | A28NLS, IMPORTIN SUBUNIT ALPHA-2 | | Authors: | Chang, C.-W, Counago, R.M, Williams, S.J, Kobe, B. | | Deposit date: | 2013-01-10 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Distinctive Conformation of Minor Site-Specific Nuclear Localization Signals Bound to Importin-Alpha
Traffic, 14, 2013
|
|
3ZIR
 
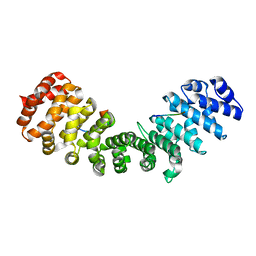 | | minor-site specific NLS (B141) | | Descriptor: | B141NLS, IMPORTIN SUBUNIT ALPHA-2 | | Authors: | Chang, C.-W, Counago, R.M, Williams, S.J, Kobe, B. | | Deposit date: | 2013-01-10 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Distinctive Conformation of Minor Site-Specific Nuclear Localization Signals Bound to Importin-Alpha
Traffic, 14, 2013
|
|
3ZIN
 
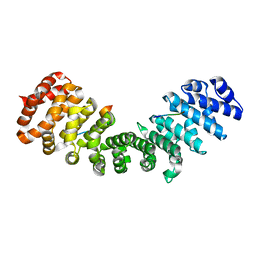 | | Gu_alpha_helicase | | Descriptor: | IMPORTIN SUBUNIT ALPHA-2, NUCLEOLAR RNA HELICASE 2 | | Authors: | Chang, C.-W, Counago, R.M, Williams, S.J, Kobe, B. | | Deposit date: | 2013-01-10 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Distinctive Conformation of Minor Site-Specific Nuclear Localization Signals Bound to Importin-Alpha
Traffic, 14, 2013
|
|
6F92
 
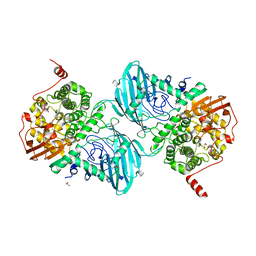 | | Structure of the family GH92 alpha-mannosidase BT3965 from Bacteroides thetaiotaomicron in complex with Mannoimidazole (ManI) | | Descriptor: | (5R,6R,7S,8R)-5-(HYDROXYMETHYL)-5,6,7,8-TETRAHYDROIMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Thompson, A.J, Spears, R.J, Zhu, Y, Suits, M.D.L, Williams, S.J, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2017-12-13 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Bacteroides thetaiotaomicron generates diverse alpha-mannosidase activities through subtle evolution of a distal substrate-binding motif.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6FAR
 
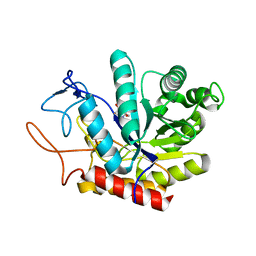 | | Structure of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with mannose-alpha-1,3-mannoimidazole | | Descriptor: | (5R,6R,7S,8R)-5-(HYDROXYMETHYL)-5,6,7,8-TETRAHYDROIMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, Glycosyl hydrolase family 71, alpha-D-mannopyranose | | Authors: | Fernandes, P.Z, Petricevic, M, Sobala, L.F, Davies, G.J, Williams, S.J. | | Deposit date: | 2017-12-16 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Exploration of Strategies for Mechanism-Based Inhibitor Design for Family GH99 endo-alpha-1,2-Mannanases.
Chemistry, 24, 2018
|
|
6F8Z
 
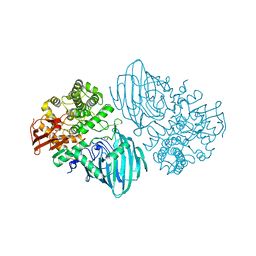 | | Structure of the family GH92 alpha-mannosidase BT3130 from Bacteroides thetaiotaomicron | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,2-mannosidase, putative, ... | | Authors: | Thompson, A.J, Spears, R.J, Zhu, Y, Suits, M.D.L, Williams, S.J, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2017-12-13 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Bacteroides thetaiotaomicron generates diverse alpha-mannosidase activities through subtle evolution of a distal substrate-binding motif.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
4AYP
 
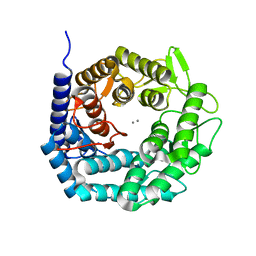 | | Structure of The GH47 processing alpha-1,2-mannosidase from Caulobacter strain K31 in complex with thiomannobioside | | Descriptor: | CALCIUM ION, MANNOSYL-OLIGOSACCHARIDE 1,2-ALPHA-MANNOSIDASE, SODIUM ION, ... | | Authors: | Thompson, A.J, Dabin, J, Iglesias-Fernandez, J, Iglesias-Fernandez, A, Dinev, Z, Williams, S.J, Siriwardena, A, Moreland, C, Hu, T.C, Smith, D.K, Gilbert, H.J, Rovira, C, Davies, G.J. | | Deposit date: | 2012-06-21 | | Release date: | 2013-01-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | The Reaction Coordinate of a Bacterial Gh47 Alpha-Mannosidase: A Combined Quantum Mechanical and Structural Approach.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
4AYR
 
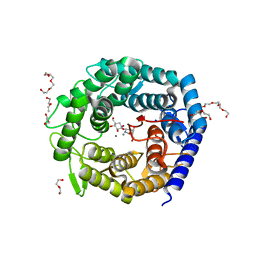 | | Structure of The GH47 processing alpha-1,2-mannosidase from Caulobacter strain K31 in complex with noeuromycin | | Descriptor: | (3S,4R,5R)-3,4-DIHYDROXY-5-(HYDROXYMETHYL)PIPERIDIN-2-ONE, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, CALCIUM ION, ... | | Authors: | Thompson, A.J, Dabin, J, Iglesias-Fernandez, J, Iglesias-Fernandez, A, Dinev, Z, Williams, S.J, Siriwardena, A, Moreland, C, Hu, T.C, Smith, D.K, Gilbert, H.J, Rovira, C, Davies, G.J. | | Deposit date: | 2012-06-21 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The Reaction Coordinate of a Bacterial Gh47 Alpha-Mannosidase: A Combined Quantum Mechanical and Structural Approach.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
5TEC
 
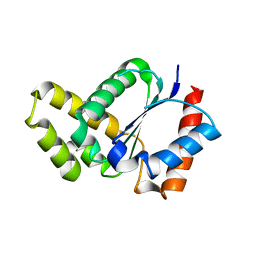 | | Crystal structure of the TIR domain from the Arabidopsis thaliana NLR protein SNC1 | | Descriptor: | Protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1 | | Authors: | Zhang, X, Bentham, A, Ve, T, Williams, S.J, Kobe, B. | | Deposit date: | 2016-09-20 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Multiple functional self-association interfaces in plant TIR domains.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4BJM
 
 | | Crystal structure of the flax-rust effector avrM | | Descriptor: | AVRM, CHLORIDE ION | | Authors: | Ve, T, Williams, S.J, Kobe, B. | | Deposit date: | 2013-04-19 | | Release date: | 2013-10-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of the Flax-Rust Effector Avrm Reveal Insights Into the Molecular Basis of Plant-Cell Entry and Effector-Triggered Immunity
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4BJN
 
 | | Crystal structure of the flax-rust effector AvrM-A | | Descriptor: | AVRM-A | | Authors: | Ve, T, Williams, S.J, Kobe, B. | | Deposit date: | 2013-04-19 | | Release date: | 2013-10-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of the Flax-Rust Effector Avrm Reveal Insights Into the Molecular Basis of Plant-Cell Entry and Effector-Triggered Immunity
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5V4L
 
 | |
4AYO
 
 | | Structure of The GH47 processing alpha-1,2-mannosidase from Caulobacter strain K31 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, MANNOSYL-OLIGOSACCHARIDE 1,2-ALPHA-MANNOSIDASE, ... | | Authors: | Thompson, A.J, Dabin, J, Iglesias-Fernandez, J, Iglesias-Fernandez, A, Dinev, Z, Williams, S.J, Siriwardena, A, Moreland, C, Hu, T.C, Smith, D.K, Gilbert, H.J, Rovira, C, Davies, G.J. | | Deposit date: | 2012-06-21 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | The Reaction Coordinate of a Bacterial Gh47 Alpha-Mannosidase: A Combined Quantum Mechanical and Structural Approach.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
4BA3
 
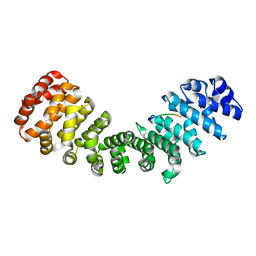 | | mImp_alphadIBB_A89NLS | | Descriptor: | A89NLS, IMPORTIN SUBUNIT ALPHA-2 | | Authors: | Chang, C.-W, Counago, R.L.M, Williams, S.J, Boden, M, Kobe, B. | | Deposit date: | 2012-09-11 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Rice Importin-Alpha and Structural Basis of its Interaction with Plant-Specific Nuclear Localization Signals.
Plant Cell, 24, 2012
|
|
4BQK
 
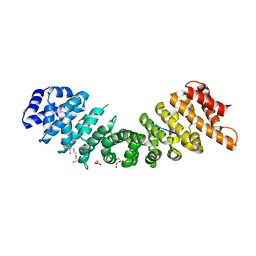 | | rice importin_alpha : VirD2NLS complex | | Descriptor: | DI(HYDROXYETHYL)ETHER, IMPORTIN SUBUNIT ALPHA-1A, T-DNA BORDER ENDONUCLEASE VIRD2 | | Authors: | Chang, C.-W, Counago, R.M, Williams, S.J, Kobe, B. | | Deposit date: | 2013-05-31 | | Release date: | 2014-02-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structural Basis of Interaction of Bipartite Nuclear Localization Signal from Agrobacterium Vird2 with Rice Importin-Alpha
Mol.Plant, 7, 2014
|
|
4BPL
 
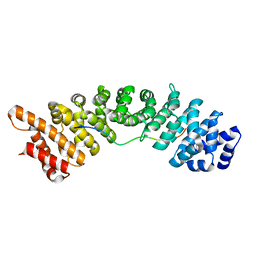 | | rice importin_alpha in complex with nucleoplasmin NLS | | Descriptor: | IMPORTIN SUBUNIT ALPHA-1A, NUCLEOPLASMIN NLS | | Authors: | Chang, C.-W, Counago, R.M, Williams, S.J, Kobe, B. | | Deposit date: | 2013-05-27 | | Release date: | 2014-02-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Interaction of Bipartite Nuclear Localization Signal from Agrobacterium Vird2 with Rice Importin-Alpha
Mol.Plant, 7, 2014
|
|
4D4B
 
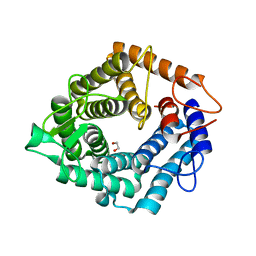 | | The catalytic domain, BcGH76, of Bacillus circulans Aman6 in complex with MSMSMe | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-1,6-MANNANASE, alpha-D-mannopyranose-(1-6)-methyl 1,6-dithio-alpha-D-mannopyranoside | | Authors: | Thompson, A.J, Speciale, G, Iglesias-Fernandez, J, Hakki, Z, Belz, T, Cartmell, A, Spears, R.J, Stepper, J, Gilbert, H.J, Rovira, C, Williams, S.J, Davies, G.J. | | Deposit date: | 2014-10-27 | | Release date: | 2015-03-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Evidence for a Boat Conformation at the Transition State of Gh76 Alpha-1,6-Mannanases- Key Enzymes in Bacterial and Fungal Mannoprotein Metabolism
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4D4C
 
 | | The catalytic domain, BcGH76, of Bacillus circulans Aman6 in complex with 1,6-ManDMJ | | Descriptor: | 1,2-ETHANEDIOL, 1-DEOXYMANNOJIRIMYCIN, ALPHA-1,6-MANNANASE, ... | | Authors: | Thompson, A.J, Speciale, G, Iglesias-Fernandez, J, Hakki, Z, Belz, T, Cartmell, A, Spears, R.J, Stepper, J, Gilbert, H.J, Rovira, C, Williams, S.J, Davies, G.J. | | Deposit date: | 2014-10-27 | | Release date: | 2015-03-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Evidence for a Boat Conformation at the Transition State of Gh76 Alpha-1,6-Mannanases- Key Enzymes in Bacterial and Fungal Mannoprotein Metabolism
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
