1E2X
 
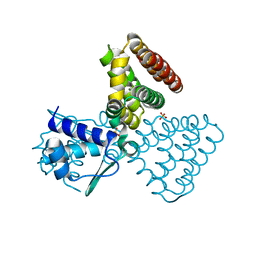 | | FadR, fatty acid responsive transcription factor from E. coli | | Descriptor: | FATTY ACID METABOLISM REGULATOR PROTEIN, SULFATE ION | | Authors: | Van Aalten, D.M.F, Dirusso, C.C, Knudsen, J, Wierenga, R.K. | | Deposit date: | 2000-05-30 | | Release date: | 2000-12-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Fadr, a Fatty Acid-Responsive Transcription Factor with a Novel Acyl Coenzyme A-Binding Fold
Embo J., 19, 2000
|
|
4QO6
 
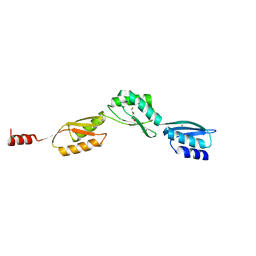 | |
5BZ4
 
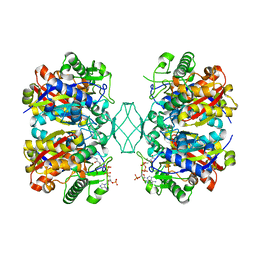 | | Crystal structure of a T1-like thiolase (CoA-complex) from Mycobacterium smegmatis | | Descriptor: | Beta-ketothiolase, COENZYME A | | Authors: | Janardan, N, Harijan, R.K, Kiema, T.R, Wierenga, R.K, Murthy, M.R.N. | | Deposit date: | 2015-06-11 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural characterization of a mitochondrial 3-ketoacyl-CoA (T1)-like thiolase from Mycobacterium smegmatis
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6YSW
 
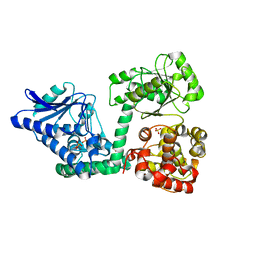 | | E. coli anaerobic trifunctional enzyme subunit-alpha in complex with coenzyme A | | Descriptor: | COENZYME A, Fatty acid oxidation complex subunit alpha, SULFATE ION | | Authors: | Sah-Teli, S.K, Hynonen, M.J, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2020-04-23 | | Release date: | 2021-05-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Structural basis for different membrane-binding properties of E. coli anaerobic and human mitochondrial beta-oxidation trifunctional enzymes
Structure, 2023
|
|
7R5T
 
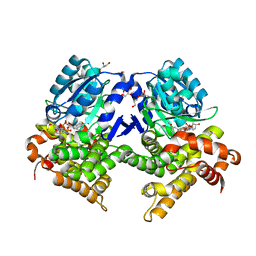 | | CRYSTAL STRUCTURE OF E.coli ALCOHOL DEHYDROGENASE - FucO MUTANT F254I COMPLEXED WITH FE, NADH, AND GLYCEROL | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ADENOSINE-5-DIPHOSPHORIBOSE, FE (III) ION, ... | | Authors: | Sridhar, S, Kiema, T.R, Wierenga, R, Widersten, M. | | Deposit date: | 2022-02-11 | | Release date: | 2022-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures of lactaldehyde reductase, FucO, link enzyme activity to hydrogen bond networks and conformational dynamics.
Febs J., 290, 2023
|
|
6YSV
 
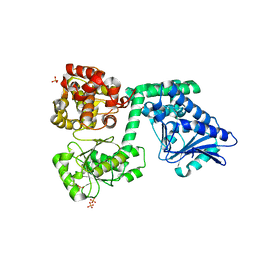 | |
6EVO
 
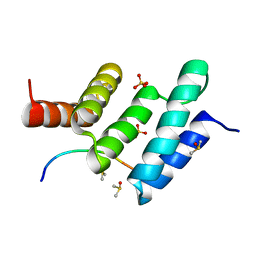 | | Crystal structure the peptide-substrate-binding domain of human type II collagen prolyl 4-hydroxylase complexed with Pro-Pro-Gly-Pro-Arg-Gly-Pro-Pro-Gly. | | Descriptor: | DIMETHYL SULFOXIDE, PRO-PRO-GLY-PRO-ARG-GLY-PRO-PRO-GLY, Prolyl 4-hydroxylase subunit alpha-2, ... | | Authors: | Murthy, A.V, Sulu, R, Koski, M.K, Wierenga, R.K. | | Deposit date: | 2017-11-02 | | Release date: | 2018-09-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural enzymology binding studies of the peptide-substrate-binding domain of human collagen prolyl 4-hydroxylase (type-II): High affinity peptides have a PxGP sequence motif.
Protein Sci., 27, 2018
|
|
6EVM
 
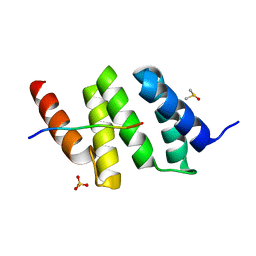 | | Crystal structure of a Pro-9 complexed peptide-substrate-binding domain of human type II collagen prolyl 4-hydroxylase | | Descriptor: | DIMETHYL SULFOXIDE, Pro-9, Prolyl 4-hydroxylase subunit alpha-2, ... | | Authors: | Murthy, A.V, Sulu, R, Koski, M.K, Wierenga, R.K. | | Deposit date: | 2017-11-02 | | Release date: | 2018-09-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural enzymology binding studies of the peptide-substrate-binding domain of human collagen prolyl 4-hydroxylase (type-II): High affinity peptides have a PxGP sequence motif.
Protein Sci., 27, 2018
|
|
6EVN
 
 | | Crystal structure of peptide-substrate-binding domain of human type II collagen prolyl 4-hydroxylase complex with Pro-Pro-Gly-Pro-Ala-Gly-Pro-Pro-Gly. | | Descriptor: | DIMETHYL SULFOXIDE, PRO-PRO-GLY-PRO-ALA-GLY-PRO-PRO-GLY, Prolyl 4-hydroxylase subunit alpha-2, ... | | Authors: | Murthy, A.V, Sulu, R, Koski, M.K, Wierenga, R.K. | | Deposit date: | 2017-11-02 | | Release date: | 2018-09-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural enzymology binding studies of the peptide-substrate-binding domain of human collagen prolyl 4-hydroxylase (type-II): High affinity peptides have a PxGP sequence motif.
Protein Sci., 27, 2018
|
|
6EVL
 
 | | Crystal structure of an unlignaded peptide-substrate-binding domain of human type II collagen prolyl 4-hydroxylase | | Descriptor: | DIMETHYL SULFOXIDE, GLYCINE, Prolyl 4-hydroxylase subunit alpha-2, ... | | Authors: | Murthy, A.V, Sulu, R, Koski, M.K, Wierenga, R.K. | | Deposit date: | 2017-11-02 | | Release date: | 2018-09-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural enzymology binding studies of the peptide-substrate-binding domain of human collagen prolyl 4-hydroxylase (type-II): High affinity peptides have a PxGP sequence motif.
Protein Sci., 27, 2018
|
|
6EVP
 
 | | Crystal structure the peptide-substrate-binding domain of human type II collagen prolyl 4-hydroxylase complexed with Pro-Pro-Gly-Pro-Glu-Gly-Pro-Pro-Gly. | | Descriptor: | DIMETHYL SULFOXIDE, PRO-PRO-GLY-PRO-GLU-GLY-PRO-PRO-GLY, Prolyl 4-hydroxylase subunit alpha-2, ... | | Authors: | Murthy, A.V, Sulu, R, Koski, M.K, Wierenga, R.K. | | Deposit date: | 2017-11-02 | | Release date: | 2018-09-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural enzymology binding studies of the peptide-substrate-binding domain of human collagen prolyl 4-hydroxylase (type-II): High affinity peptides have a PxGP sequence motif.
Protein Sci., 27, 2018
|
|
7QH0
 
 | |
4ZRC
 
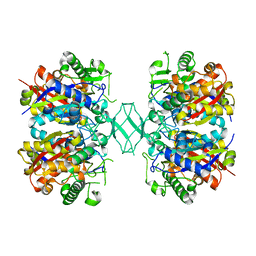 | | Crystal structure of MSM-13, a putative T1-like thiolase from Mycobacterium smegmatis | | Descriptor: | Beta-ketothiolase | | Authors: | Janardan, N, Harijan, R.K, Keima, T.R, Wierenga, R, Murthy, M.R.N. | | Deposit date: | 2015-05-12 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural characterization of a mitochondrial 3-ketoacyl-CoA (T1)-like thiolase from Mycobacterium smegmatis
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7O1K
 
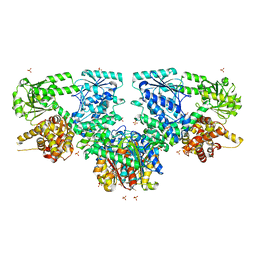 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme alpha-E141A, beta-C92A mutant | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, GLYCEROL, Putative acyltransferase Rv0859, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2021-03-29 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Substrate specificity and conformational flexibility properties of the Mycobacterium tuberculosis beta-oxidation trifunctional enzyme.
J.Struct.Biol., 213, 2021
|
|
7O4V
 
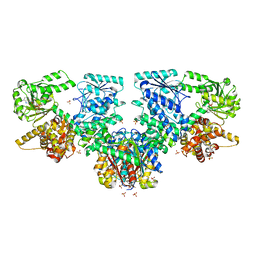 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme in complex with oxidized nicotinamide adenine dinucleotide | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative acyltransferase Rv0859, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2021-04-07 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Substrate specificity and conformational flexibility properties of the Mycobacterium tuberculosis beta-oxidation trifunctional enzyme.
J.Struct.Biol., 213, 2021
|
|
7O4Q
 
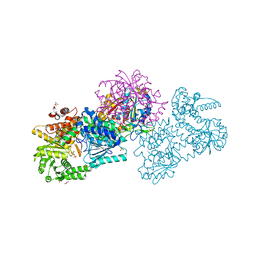 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme in space group C2221 (unliganded) | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, GLYCEROL, Putative acyltransferase Rv0859, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2021-04-07 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Substrate specificity and conformational flexibility properties of the Mycobacterium tuberculosis beta-oxidation trifunctional enzyme.
J.Struct.Biol., 213, 2021
|
|
7O4T
 
 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme with Coenzyme A bound at the hydratase, thiolase active sites and possible additional binding site (CoA(ECH/HAD)) | | Descriptor: | 3'-PHOSPHATE-ADENOSINE-5'-DIPHOSPHATE, 3-hydroxyacyl-CoA dehydrogenase, COENZYME A, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2021-04-07 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Substrate specificity and conformational flexibility properties of the Mycobacterium tuberculosis beta-oxidation trifunctional enzyme.
J.Struct.Biol., 213, 2021
|
|
7O4S
 
 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme with Coenzyme A bound at the hydratase, thiolase active sites and additional binding site (CoA(ECH2)) | | Descriptor: | 3'-PHOSPHATE-ADENOSINE-5'-DIPHOSPHATE, 3-hydroxyacyl-CoA dehydrogenase, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2021-04-07 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Substrate specificity and conformational flexibility properties of the Mycobacterium tuberculosis beta-oxidation trifunctional enzyme.
J.Struct.Biol., 213, 2021
|
|
7O1J
 
 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme beta-C92A mutant | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, GLYCEROL, Putative acyltransferase Rv0859, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2021-03-29 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Substrate specificity and conformational flexibility properties of the Mycobacterium tuberculosis beta-oxidation trifunctional enzyme.
J.Struct.Biol., 213, 2021
|
|
7O4R
 
 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme with Coenzyme A bound at the thiolase active sites and additional binding site (CoA(HAD/KAT)) | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, COENZYME A, GLYCEROL, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2021-04-07 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Substrate specificity and conformational flexibility properties of the Mycobacterium tuberculosis beta-oxidation trifunctional enzyme.
J.Struct.Biol., 213, 2021
|
|
1DCI
 
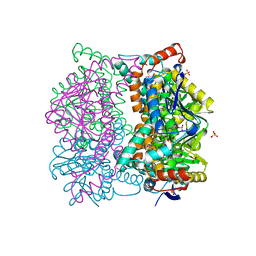 | | DIENOYL-COA ISOMERASE | | Descriptor: | 1,2-ETHANEDIOL, DIENOYL-COA ISOMERASE, MAGNESIUM ION, ... | | Authors: | Modis, Y, Filppula, S.A, Novikov, D, Norledge, B, Hiltunen, J.K, Wierenga, R.K. | | Deposit date: | 1998-02-13 | | Release date: | 1999-03-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of dienoyl-CoA isomerase at 1.5 A resolution reveals the importance of aspartate and glutamate sidechains for catalysis.
Structure, 6, 1998
|
|
7O1I
 
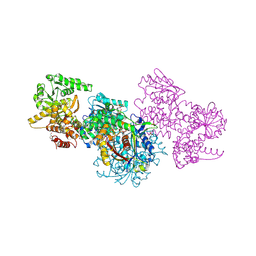 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme alpha-E141A mutant | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, COENZYME A, GLYCEROL, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2021-03-29 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Substrate specificity and conformational flexibility properties of the Mycobacterium tuberculosis beta-oxidation trifunctional enzyme.
J.Struct.Biol., 213, 2021
|
|
7O1G
 
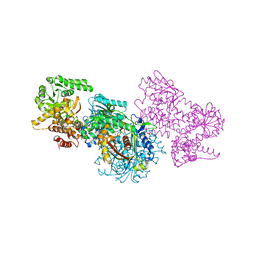 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme alpha-E141A-H462A, beta-C92A mutant | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, Putative acyltransferase Rv0859, SULFATE ION | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2021-03-29 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Substrate specificity and conformational flexibility properties of the Mycobacterium tuberculosis beta-oxidation trifunctional enzyme.
J.Struct.Biol., 213, 2021
|
|
7O1L
 
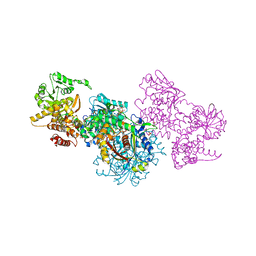 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme alpha-H462A mutant | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, COENZYME A, GLYCEROL, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2021-03-29 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Substrate specificity and conformational flexibility properties of the Mycobacterium tuberculosis beta-oxidation trifunctional enzyme.
J.Struct.Biol., 213, 2021
|
|
7O1M
 
 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme alpha-H462A, beta-C92A mutant | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, GLYCEROL, Putative acyltransferase Rv0859, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2021-03-29 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Substrate specificity and conformational flexibility properties of the Mycobacterium tuberculosis beta-oxidation trifunctional enzyme.
J.Struct.Biol., 213, 2021
|
|
