3ZBK
 
 | | Crystal structure of SCP2 thiolase from Leishmania mexicana: The C123A mutant. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-KETOACYL-COA THIOLASE-LIKE PROTEIN, CHLORIDE ION, ... | | Authors: | Harijan, R.K, Kiema, T.-R, Weiss, M.S, Michels, P.A.M, Wierenga, R.K. | | Deposit date: | 2012-11-10 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of Scp2-Thiolases of Trypanosomatidae, Human Pathogens Causing Widespread Tropical Diseases: The Importance for Catalysis of the Cysteine of the Unique Hdcf Loop.
Biochem.J., 455, 2013
|
|
3PH9
 
 | | Crystal structure of the human anterior gradient protein 3 | | Descriptor: | Anterior gradient protein 3 homolog | | Authors: | Nguyen, V.D, Ruddock, L.W, Salin, M, Wierenga, R.K. | | Deposit date: | 2010-11-03 | | Release date: | 2011-10-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of human anterior gradient protein 3.
Acta Crystallogr.,Sect.F, 74, 2018
|
|
5LNQ
 
 | |
3GZE
 
 | |
6TP5
 
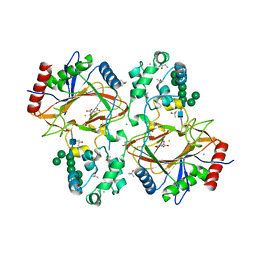 | | Crystal structure of human Transmembrane prolyl 4-hydroxylase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Myllykoski, M, Sutinen, A, Koski, M.K, Kallio, J.P, Raasakka, A, Myllyharju, J, Wierenga, R.K, Koivunen, P. | | Deposit date: | 2019-12-12 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of transmembrane prolyl 4-hydroxylase reveals unique organization of EF and dioxygenase domains.
J.Biol.Chem., 296, 2020
|
|
2DUB
 
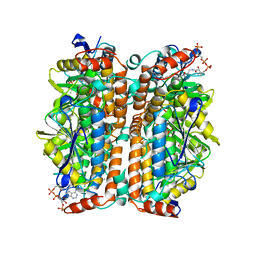 | | ENOYL-COA HYDRATASE COMPLEXED WITH OCTANOYL-COA | | Descriptor: | 2-ENOYL-COA HYDRATASE, OCTANOYL-COENZYME A | | Authors: | Engel, C.K, Wierenga, R.K. | | Deposit date: | 1997-04-28 | | Release date: | 1998-04-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of enoyl-CoA hydratase complexed with octanoyl-CoA reveals the structural adaptations required for binding of a long chain fatty acid-CoA molecule.
J.Mol.Biol., 275, 1998
|
|
4U1A
 
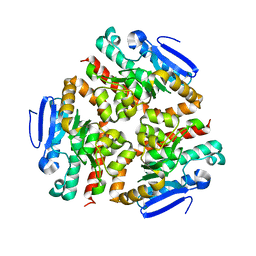 | | Crystal structure of human peroxisomal delta3,delta2, enoyl-CoA isomerase helix-10 deletion mutant (ISOB-ECI2) | | Descriptor: | CHLORIDE ION, Enoyl-CoA delta isomerase 2 | | Authors: | Onwukwe, G.U, Koski, M.K, Wierenga, R.K. | | Deposit date: | 2014-07-15 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Human Delta (3) , Delta (2) -enoyl-CoA isomerase, type 2: a structural enzymology study on the catalytic role of its ACBP domain and helix-10.
Febs J., 282, 2015
|
|
4U18
 
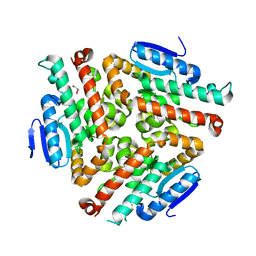 | | Crystal structure of human peroxisomal delta3,delta2, enoyl-CoA isomerase (ISO-ECI2) | | Descriptor: | CHLORIDE ION, Enoyl-CoA delta isomerase 2, mitochondrial, ... | | Authors: | Onwukwe, G.U, Koski, M.K, Wierenga, R.K. | | Deposit date: | 2014-07-15 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Human Delta (3) , Delta (2) -enoyl-CoA isomerase, type 2: a structural enzymology study on the catalytic role of its ACBP domain and helix-10.
Febs J., 282, 2015
|
|
4U19
 
 | | Crystal structure of human peroxisomal delta3,delta2, enoyl-CoA isomerase V349A mutant (ISOA-ECI2) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Enoyl-CoA delta isomerase 2 | | Authors: | Onwukwe, G.U, Koski, M.K, Wierenga, R.K. | | Deposit date: | 2014-07-15 | | Release date: | 2014-12-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Human Delta (3) , Delta (2) -enoyl-CoA isomerase, type 2: a structural enzymology study on the catalytic role of its ACBP domain and helix-10.
Febs J., 282, 2015
|
|
5F0V
 
 | | X-ray crystal structure of a thiolase from Escherichia coli at 1.8 A resolution | | Descriptor: | 1,2-ETHANEDIOL, Acetyl-CoA acetyltransferase | | Authors: | Ithayaraja, M, Neelanjana, J, Wierenga, R, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2015-11-28 | | Release date: | 2016-07-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a thiolase from Escherichia coli at 1.8 angstrom resolution.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5F38
 
 | | X-ray crystal structure of a thiolase from Escherichia coli at 1.8 A resolution | | Descriptor: | 1,2-ETHANEDIOL, Acetyl-CoA acetyltransferase, COENZYME A, ... | | Authors: | Ithayaraja, M, Neelanjana, J, Wierenga, R, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2015-12-02 | | Release date: | 2016-07-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a thiolase from Escherichia coli at 1.8 angstrom resolution.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5BYV
 
 | | Crystal structure of MSM-13, a putative T1-like thiolase from Mycobacterium smegmatis | | Descriptor: | Beta-ketothiolase | | Authors: | Janardan, N, Harijan, R.K, Keima, T.R, Wierenga, R, Murthy, M.R.N. | | Deposit date: | 2015-06-11 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.162 Å) | | Cite: | Structural characterization of a mitochondrial 3-ketoacyl-CoA (T1)-like thiolase from Mycobacterium smegmatis
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2CB8
 
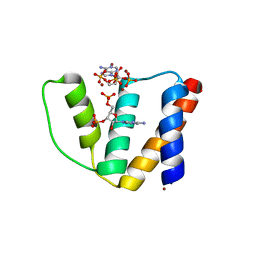 | | High resolution crystal structure of liganded human L-ACBP | | Descriptor: | 2-METHOXYETHANOL, ACYL-COA-BINDING PROTEIN, SULFATE ION, ... | | Authors: | Taskinen, J.P, van Aalten, D.M, Knudsen, J, Wierenga, R.K. | | Deposit date: | 2006-01-03 | | Release date: | 2006-10-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High Resolution Crystal Structures of Unliganded and Liganded Human Liver Acbp Reveal a New Mode of Binding for the Acyl-Coa Ligand.
Proteins: Struct., Funct., Bioinf., 66, 2007
|
|
7AYC
 
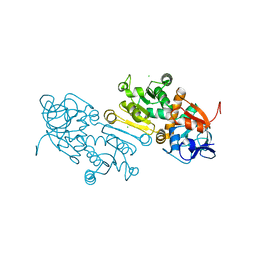 | | Crystal Structure of human mitochondrial 2-Enoyl Thioester Reductase (MECR) with single mutation G165Q | | Descriptor: | CHLORIDE ION, Enoyl-[acyl-carrier-protein] reductase, mitochondrial | | Authors: | Rahman, M.T, Koski, M.K, Autio, K.J, Kastaniotis, A.J, Wierenga, R.K, Hiltunen, J.K. | | Deposit date: | 2020-11-12 | | Release date: | 2022-06-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | An engineered variant of MECR reductase reveals indispensability of long-chain acyl-ACPs for mitochondrial respiration.
Nat Commun, 14, 2023
|
|
7AYB
 
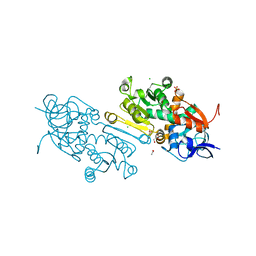 | | Crystal Structure of wild type human mitochondrial 2-Enoyl Thioester Reductase (MECR) | | Descriptor: | ACETIC ACID, CHLORIDE ION, Enoyl-[acyl-carrier-protein] reductase, ... | | Authors: | Rahman, M.T, Koski, M.K, Autio, K.J, Kastaniotis, A.J, Wierenga, R.K, Hiltunen, J.K. | | Deposit date: | 2020-11-12 | | Release date: | 2022-06-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | An engineered variant of MECR reductase reveals indispensability of long-chain acyl-ACPs for mitochondrial respiration.
Nat Commun, 14, 2023
|
|
1WL4
 
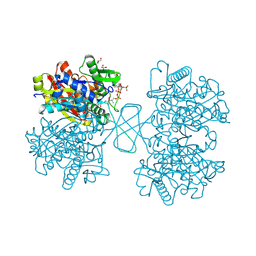 | | Human cytosolic acetoacetyl-CoA thiolase complexed with CoA | | Descriptor: | COENZYME A, GLYCEROL, SULFATE ION, ... | | Authors: | Kursula, P, Fukao, T, Kondo, N, Wierenga, R.K. | | Deposit date: | 2004-06-20 | | Release date: | 2005-03-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | High Resolution Crystal Structures of Human Cytosolic Thiolase (CT): A Comparison of the Active Sites of Human CT, Bacterial Thiolase, and Bacterial KAS I
J.Mol.Biol., 347, 2005
|
|
1WL5
 
 | | Human cytosolic acetoacetyl-CoA thiolase | | Descriptor: | GLYCEROL, SULFATE ION, acetyl-Coenzyme A acetyltransferase 2 | | Authors: | Kursula, P, Fukao, T, Kondo, N, Wierenga, R.K. | | Deposit date: | 2004-06-20 | | Release date: | 2005-03-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | High Resolution Crystal Structures of Human Cytosolic Thiolase (CT): A Comparison of the Active Sites of Human CT, Bacterial Thiolase, and Bacterial KAS I
J.Mol.Biol., 347, 2005
|
|
1X74
 
 | | Alpha-methylacyl-CoA racemase from Mycobacterium tuberculosis- mutational and structural characterization of the fold and active site | | Descriptor: | 2-methylacyl-CoA racemase, GLYCEROL, PHOSPHATE ION | | Authors: | Kalle, S, Bhaumik, P, Schmitz, W, Kotti, T.J, Conzelmann, E, Wierenga, R.K, Hiltunen, J.K. | | Deposit date: | 2004-08-13 | | Release date: | 2005-01-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | {alpha}-Methylacyl-CoA Racemase from Mycobacterium tuberculosis: MUTATIONAL AND STRUCTURAL CHARACTERIZATION OF THE ACTIVE SITE AND THE FOLD
J.Biol.Chem., 280, 2005
|
|
1ZCJ
 
 | |
6EVO
 
 | | Crystal structure the peptide-substrate-binding domain of human type II collagen prolyl 4-hydroxylase complexed with Pro-Pro-Gly-Pro-Arg-Gly-Pro-Pro-Gly. | | Descriptor: | DIMETHYL SULFOXIDE, PRO-PRO-GLY-PRO-ARG-GLY-PRO-PRO-GLY, Prolyl 4-hydroxylase subunit alpha-2, ... | | Authors: | Murthy, A.V, Sulu, R, Koski, M.K, Wierenga, R.K. | | Deposit date: | 2017-11-02 | | Release date: | 2018-09-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural enzymology binding studies of the peptide-substrate-binding domain of human collagen prolyl 4-hydroxylase (type-II): High affinity peptides have a PxGP sequence motif.
Protein Sci., 27, 2018
|
|
5BZ4
 
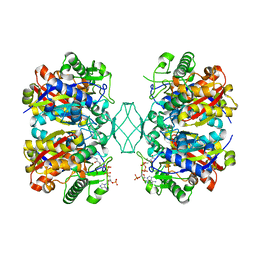 | | Crystal structure of a T1-like thiolase (CoA-complex) from Mycobacterium smegmatis | | Descriptor: | Beta-ketothiolase, COENZYME A | | Authors: | Janardan, N, Harijan, R.K, Kiema, T.R, Wierenga, R.K, Murthy, M.R.N. | | Deposit date: | 2015-06-11 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural characterization of a mitochondrial 3-ketoacyl-CoA (T1)-like thiolase from Mycobacterium smegmatis
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6EVM
 
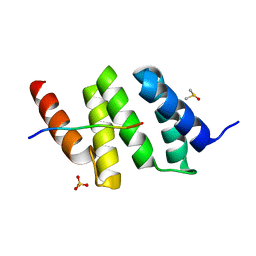 | | Crystal structure of a Pro-9 complexed peptide-substrate-binding domain of human type II collagen prolyl 4-hydroxylase | | Descriptor: | DIMETHYL SULFOXIDE, Pro-9, Prolyl 4-hydroxylase subunit alpha-2, ... | | Authors: | Murthy, A.V, Sulu, R, Koski, M.K, Wierenga, R.K. | | Deposit date: | 2017-11-02 | | Release date: | 2018-09-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural enzymology binding studies of the peptide-substrate-binding domain of human collagen prolyl 4-hydroxylase (type-II): High affinity peptides have a PxGP sequence motif.
Protein Sci., 27, 2018
|
|
6EVN
 
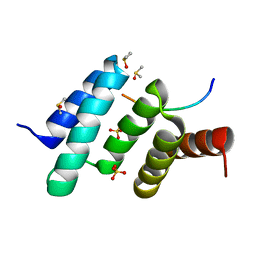 | | Crystal structure of peptide-substrate-binding domain of human type II collagen prolyl 4-hydroxylase complex with Pro-Pro-Gly-Pro-Ala-Gly-Pro-Pro-Gly. | | Descriptor: | DIMETHYL SULFOXIDE, PRO-PRO-GLY-PRO-ALA-GLY-PRO-PRO-GLY, Prolyl 4-hydroxylase subunit alpha-2, ... | | Authors: | Murthy, A.V, Sulu, R, Koski, M.K, Wierenga, R.K. | | Deposit date: | 2017-11-02 | | Release date: | 2018-09-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural enzymology binding studies of the peptide-substrate-binding domain of human collagen prolyl 4-hydroxylase (type-II): High affinity peptides have a PxGP sequence motif.
Protein Sci., 27, 2018
|
|
6EVL
 
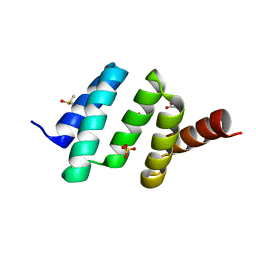 | | Crystal structure of an unlignaded peptide-substrate-binding domain of human type II collagen prolyl 4-hydroxylase | | Descriptor: | DIMETHYL SULFOXIDE, GLYCINE, Prolyl 4-hydroxylase subunit alpha-2, ... | | Authors: | Murthy, A.V, Sulu, R, Koski, M.K, Wierenga, R.K. | | Deposit date: | 2017-11-02 | | Release date: | 2018-09-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural enzymology binding studies of the peptide-substrate-binding domain of human collagen prolyl 4-hydroxylase (type-II): High affinity peptides have a PxGP sequence motif.
Protein Sci., 27, 2018
|
|
6EVP
 
 | | Crystal structure the peptide-substrate-binding domain of human type II collagen prolyl 4-hydroxylase complexed with Pro-Pro-Gly-Pro-Glu-Gly-Pro-Pro-Gly. | | Descriptor: | DIMETHYL SULFOXIDE, PRO-PRO-GLY-PRO-GLU-GLY-PRO-PRO-GLY, Prolyl 4-hydroxylase subunit alpha-2, ... | | Authors: | Murthy, A.V, Sulu, R, Koski, M.K, Wierenga, R.K. | | Deposit date: | 2017-11-02 | | Release date: | 2018-09-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural enzymology binding studies of the peptide-substrate-binding domain of human collagen prolyl 4-hydroxylase (type-II): High affinity peptides have a PxGP sequence motif.
Protein Sci., 27, 2018
|
|
