1CHD
 
 | | CHEB METHYLESTERASE DOMAIN | | Descriptor: | CHEB METHYLESTERASE | | Authors: | West, A.H, Martinez-Hackert, E, Stock, A.M. | | Deposit date: | 1995-03-09 | | Release date: | 1996-01-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the catalytic domain of the chemotaxis receptor methylesterase, CheB.
J.Mol.Biol., 250, 1995
|
|
6M7W
 
 | |
5KBX
 
 | |
5J62
 
 | | FMN-dependent Nitroreductase (CDR20291_0684) from Clostridium difficile R20291 | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Wang, B, Powell, S.M, Hessami, N, Najar, F.Z, Thomas, L.M, West, A.H, Karr, E.A, Richter-Addo, G.B. | | Deposit date: | 2016-04-04 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures of two nitroreductases from hypervirulent Clostridium difficile and functionally related interactions with the antibiotic metronidazole.
Nitric Oxide, 60, 2016
|
|
5J6C
 
 | | FMN-dependent Nitroreductase (CDR20291_0767) from Clostridium difficile R20291 | | Descriptor: | FLAVIN MONONUCLEOTIDE, IMIDAZOLE, Putative reductase | | Authors: | Powell, S.M, Wang, B, Hessami, N, Najar, F.Z, Thomas, L.M, West, A.H, Karr, E.A, Richter-Addo, G.B. | | Deposit date: | 2016-04-04 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Crystal structures of two nitroreductases from hypervirulent Clostridium difficile and functionally related interactions with the antibiotic metronidazole.
Nitric Oxide, 60, 2016
|
|
5DYM
 
 | | Crystal structure of a PadR family transcription regulator from hypervirulent Clostridium difficile R20291 - CdPadR_0991 to 1.89 Angstrom resolution | | Descriptor: | PadR-family transcriptional regulator | | Authors: | Isom, C.E, Karr, E.A, Menon, S.K, West, A.H, Richter-Addo, G.B. | | Deposit date: | 2015-09-24 | | Release date: | 2016-10-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.894 Å) | | Cite: | Crystal structure and DNA binding activity of a PadR family transcription regulator from hypervirulent Clostridium difficile R20291.
Bmc Microbiol., 16, 2016
|
|
3UGK
 
 | | Crystal Structure of C205S mutant and Saccharopine Dehydrogenase from Saccharomyces cerevisiae. | | Descriptor: | Saccharopine dehydrogenase [NAD+, L-lysine-forming] | | Authors: | Cook, P.F, Kumar, V.P, Thomas, L.M, West, A.H, Bobyk, K.D. | | Deposit date: | 2011-11-02 | | Release date: | 2012-02-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Evidence in Support of Lysine 77 and Histidine 96 as Acid-Base Catalytic Residues in Saccharopine Dehydrogenase from Saccharomyces cerevisiae.
Biochemistry, 51, 2012
|
|
3UHA
 
 | | Crystal Structure of Saccharopine Dehydrogenase from Saccharomyces cervisiae complexed with NAD. | | Descriptor: | CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Saccharopine dehydrogenase [NAD+, ... | | Authors: | Cook, P.F, Kumar, V.P, Thomas, L.M, West, A.H, Bobyk, K.D. | | Deposit date: | 2011-11-03 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evidence in Support of Lysine 77 and Histidine 96 as Acid-Base Catalytic Residues in Saccharopine Dehydrogenase from Saccharomyces cerevisiae.
Biochemistry, 51, 2012
|
|
3UH1
 
 | | Crystal Structure of Saccharopine Dehydrogenase from Saccharomyces cerevisiae with bound saccharopine and NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GLYCEROL, N-(5-AMINO-5-CARBOXYPENTYL)GLUTAMIC ACID, ... | | Authors: | Kumar, V.P, Thomas, L.M, Bobyk, K.D, Andi, B, West, A.H, Cook, P.F. | | Deposit date: | 2011-11-03 | | Release date: | 2012-02-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Evidence in Support of Lysine 77 and Histidine 96 as Acid-Base Catalytic Residues in Saccharopine Dehydrogenase from Saccharomyces cerevisiae.
Biochemistry, 51, 2012
|
|
1OXK
 
 | |
1OXB
 
 | |
1NPG
 
 | | MYOGLOBIN (HORSE HEART) WILD-TYPE COMPLEXED WITH NITROSOETHANE | | Descriptor: | Myoglobin, NITROSOETHANE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Copeland, D.M, West, A.H, Richter-Addo, G.B. | | Deposit date: | 2003-01-17 | | Release date: | 2003-11-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of ferrous horse heart myoglobin complexed with nitric oxide and nitrosoethane.
Proteins, 53, 2003
|
|
1NPF
 
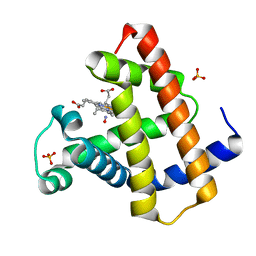 | | MYOGLOBIN (HORSE HEART) WILD-TYPE COMPLEXED WITH NITRIC OXIDE | | Descriptor: | Myoglobin, NITRIC OXIDE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Copeland, D.M, West, A.H, Richter-Addo, G.B. | | Deposit date: | 2003-01-17 | | Release date: | 2003-11-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of ferrous horse heart myoglobin complexed with nitric oxide and nitrosoethane.
Proteins, 53, 2003
|
|
5LWX
 
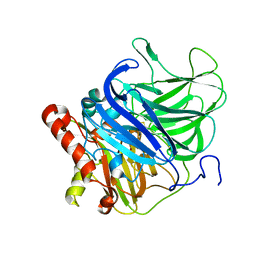 | | Crystal structure of the H253D mutant of McoG from Aspergillus niger | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | Ferraroni, M, Briganti, F, Tamayo-Ramos, J.A, van Berkel, W.J.H, Westphal, A.H. | | Deposit date: | 2016-09-19 | | Release date: | 2017-05-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structure and function of Aspergillus niger laccase McoG
Biocatalysis, 2017
|
|
1A2O
 
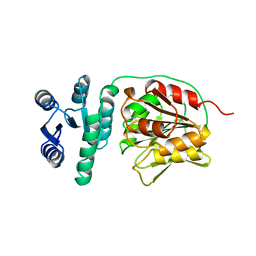 | | STRUCTURAL BASIS FOR METHYLESTERASE CHEB REGULATION BY A PHOSPHORYLATION-ACTIVATED DOMAIN | | Descriptor: | CHEB METHYLESTERASE | | Authors: | Djordjevic, S, Goudreau, P.N, Xu, Q, Stock, A.M, West, A.H. | | Deposit date: | 1998-01-06 | | Release date: | 1998-04-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for methylesterase CheB regulation by a phosphorylation-activated domain.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1QSP
 
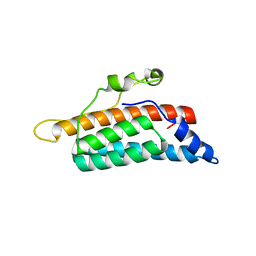 | | CRYSTAL STRUCTURE OF THE YEAST PHOSPHORELAY PROTEIN YPD1 | | Descriptor: | YPD1 | | Authors: | Xu, Q, West, A.H. | | Deposit date: | 1999-06-22 | | Release date: | 1999-10-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Conservation of structure and function among histidine-containing phosphotransfer (HPt) domains as revealed by the crystal structure of YPD1.
J.Mol.Biol., 292, 1999
|
|
2FRI
 
 | | Horse Heart Myoglobin, Nitrite Adduct, Co-crystallized | | Descriptor: | Myoglobin, NITRITE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Copeland, D.M, Soares, A.S, West, A.H, Richter-Addo, G.B. | | Deposit date: | 2006-01-19 | | Release date: | 2006-05-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of the nitrite and nitric oxide complexes of horse heart myoglobin.
J.Inorg.Biochem., 100, 2006
|
|
2FRK
 
 | | Nitrosyl Horse Heart Myoglobin, Nitric Oxide Gas Method | | Descriptor: | Myoglobin, NITRIC OXIDE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Copeland, D.M, Soares, A.S, West, A.H, Richter-Addo, G.B. | | Deposit date: | 2006-01-19 | | Release date: | 2006-05-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structures of the nitrite and nitric oxide complexes of horse heart myoglobin.
J.Inorg.Biochem., 100, 2006
|
|
2FRF
 
 | | Horse Heart Myoglobin, Nitrite Adduct, Crystal Soak | | Descriptor: | Myoglobin, NITRITE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Copeland, D.M, Soares, A.S, West, A.H, Richter-Addo, G.B. | | Deposit date: | 2006-01-19 | | Release date: | 2006-05-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structures of the nitrite and nitric oxide complexes of horse heart myoglobin.
J.Inorg.Biochem., 100, 2006
|
|
2FRJ
 
 | | Nitrosyl Horse Heart Myoglobin, Nitrite/Dithionite Method | | Descriptor: | Myoglobin, NITRIC OXIDE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Copeland, D.M, Soares, A.S, West, A.H, Richter-Addo, G.B. | | Deposit date: | 2006-01-19 | | Release date: | 2006-05-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structures of the nitrite and nitric oxide complexes of horse heart myoglobin.
J.Inorg.Biochem., 100, 2006
|
|
2QRL
 
 | | Crystal Structure of Oxalylglycine-bound Saccharopine Dehydrogenase (L-Lys Forming) from Saccharomyces cerevisiae | | Descriptor: | N-OXALYLGLYCINE, Saccharopine dehydrogenase, NAD+, ... | | Authors: | Andi, B, Xu, H, Cook, P.F, West, A.H. | | Deposit date: | 2007-07-28 | | Release date: | 2007-10-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of Ligand-Bound Saccharopine Dehydrogenase from Saccharomyces cerevisiae
Biochemistry, 46, 2007
|
|
2R25
 
 | | Complex of YPD1 and SLN1-R1 with bound Mg2+ and BeF3- | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, Osmosensing histidine protein kinase SLN1, ... | | Authors: | Copeland, D.M, Zhao, X, Soares, A.S, West, A.H. | | Deposit date: | 2007-08-24 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a complex between the phosphorelay protein YPD1 and
the response regulator domain of SLN1 bound to a phosphoryl analog
J.Mol.Biol., 375, 2008
|
|
2QRK
 
 | | Crystal Structure of AMP-bound Saccharopine Dehydrogenase (L-Lys Forming) from Saccharomyces cerevisiae | | Descriptor: | ADENOSINE MONOPHOSPHATE, Saccharopine dehydrogenase [NAD+, L-lysine-forming | | Authors: | Andi, B, Xu, H, Cook, P.F, West, A.H. | | Deposit date: | 2007-07-28 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structures of Ligand-Bound Saccharopine Dehydrogenase from Saccharomyces cerevisiae
Biochemistry, 46, 2007
|
|
2QRJ
 
 | | Crystal Structure of Sulfate-bound Saccharopine Dehydrogenase (L-Lys Forming) from Saccharomyces cerevisiae | | Descriptor: | CHLORIDE ION, SULFATE ION, Saccharopine dehydrogenase, ... | | Authors: | Andi, B, Xu, H, Cook, P.F, West, A.H. | | Deposit date: | 2007-07-28 | | Release date: | 2007-10-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of Ligand-Bound Saccharopine Dehydrogenase from Saccharomyces cerevisiae
Biochemistry, 46, 2007
|
|
2AXQ
 
 | |
