7VCM
 
 | | crystal structure of GINKO1 | | Descriptor: | Green fluorescent protein,Potassium binding protein Kbp,Green fluorescent protein, POTASSIUM ION | | Authors: | Wen, Y, Campbell, R.E, Lemieux, M.J. | | Deposit date: | 2021-09-03 | | Release date: | 2022-07-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A sensitive and specific genetically-encoded potassium ion biosensor for in vivo applications across the tree of life.
Plos Biol., 20, 2022
|
|
6KZH
 
 | |
6KZG
 
 | |
6KYX
 
 | |
8WOH
 
 | |
8Z2O
 
 | | Crystal structure of 5-N-alpha-glycinylthymidine (N-alpha-GlyT) FAD-dependent lyase gp47/NGTO from Pseudomonads phage PaMx11 | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, Flavin-dependent lyase, ... | | Authors: | Wen, Y, Guo, W.T, Wu, B.X. | | Deposit date: | 2024-04-13 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the biosynthetic mechanism of N alpha-GlyT and 5-NmdU hypermodifications of DNA.
Nucleic Acids Res., 52, 2024
|
|
8Z2M
 
 | |
8Z2N
 
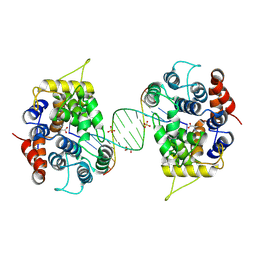 | | Crystal structure of 5-phosphomethyl-2'-deoxyuridine (5-PmdU) glycinyltransferase gp46/PUGT from Pseudomonads phage PaMx11 in complex with dsDNA | | Descriptor: | CALCIUM ION, DNA (5'-D(*TP*AP*GP*TP*CP*GP*AP*CP*GP*AP*CP*TP*A)-3'), DNA (5'-D(*TP*AP*GP*TP*CP*GP*TP*CP*GP*AP*CP*TP*A)-3'), ... | | Authors: | Wen, Y, Guo, W.T, Wu, B.X. | | Deposit date: | 2024-04-13 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the biosynthetic mechanism of N alpha-GlyT and 5-NmdU hypermodifications of DNA.
Nucleic Acids Res., 52, 2024
|
|
8Z79
 
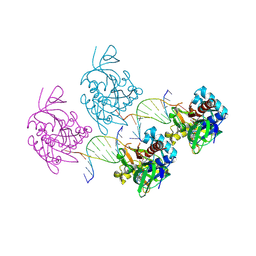 | |
7CNU
 
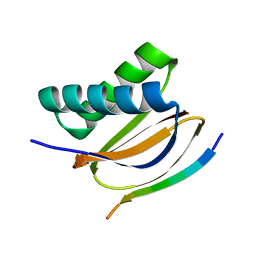 | | Crystal structure of DLC2 in complex with BMF peptide | | Descriptor: | Bcl-2-modifying factor, Dynein light chain 2, cytoplasmic | | Authors: | Wen, Y, Shao, Y. | | Deposit date: | 2020-08-03 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Non-canonical phosphorylation of Bmf by p38 MAPK promotes its apoptotic activity in anoikis.
Cell Death Differ., 29, 2022
|
|
7DMX
 
 | |
7DNB
 
 | | Crystal structure of PhoCl barrel | | Descriptor: | PhoCl Barrel, SODIUM ION | | Authors: | Wen, Y, Lemieux, J.M. | | Deposit date: | 2020-12-09 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Photocleavable proteins that undergo fast and efficient dissociation.
Chem Sci, 12, 2021
|
|
7E9Y
 
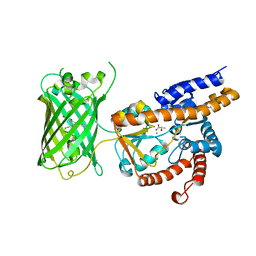 | | Crystal structure of eLACCO1 | | Descriptor: | (2S)-2-HYDROXYPROPANOIC ACID, CALCIUM ION, Lactate-binding periplasmic protein TTHA0766,Lactate-binding periplasmic protein TTHA0766 | | Authors: | Wen, Y, Campbell, R.E, Lemieux, M.J, Nasu, Y. | | Deposit date: | 2021-03-05 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A genetically encoded fluorescent biosensor for extracellular L-lactate.
Nat Commun, 12, 2021
|
|
7DNA
 
 | |
7CCI
 
 | |
7CCH
 
 | | Acinetobacter baumannii histidine kinase AdeS | | Descriptor: | AdeS | | Authors: | Wen, Y, Felix, J. | | Deposit date: | 2020-06-17 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.848 Å) | | Cite: | Proteolysis and multimerization regulate signaling along the two-component regulatory system AdeRS.
Iscience, 24, 2021
|
|
7YW0
 
 | | Bacteroides fragilis Hcp5 | | Descriptor: | Bacterodales T6SS protein TssD (Hcp) | | Authors: | Wen, Y, He, W, Bai, Y. | | Deposit date: | 2022-08-20 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure and assembly of type VI secretion system cargo delivery vehicle.
Cell Rep, 42, 2023
|
|
7DJX
 
 | | Clec4f Nanobody 246 | | Descriptor: | Clec4f Nanobody 246 | | Authors: | Wen, Y, Zheng, F. | | Deposit date: | 2020-11-22 | | Release date: | 2021-11-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Clec4f Nanobody 246
To Be Published
|
|
7DJY
 
 | | Clec4f Nanobody 322 | | Descriptor: | Clec4f Nanobody 322 | | Authors: | Wen, Y, Zheng, F. | | Deposit date: | 2020-11-22 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Clec4f Nanobody 322
To Be Published
|
|
7EYK
 
 | |
7EYP
 
 | | Crystal structure of Pseudomonas aeruginosa ppnP | | Descriptor: | Pyrimidine/purine nucleoside phosphorylase | | Authors: | Wen, Y, Wu, B.X. | | Deposit date: | 2021-05-31 | | Release date: | 2022-02-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of a new class of pyrimidine/purine nucleoside phosphorylase revealed a Cupin fold.
Proteins, 90, 2022
|
|
7EYJ
 
 | | Crystal structure of Escherichia coli ppnP | | Descriptor: | Pyrimidine/purine nucleoside phosphorylase, SULFATE ION | | Authors: | Wen, Y, Wu, B.X. | | Deposit date: | 2021-05-31 | | Release date: | 2022-02-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Crystal structures of a new class of pyrimidine/purine nucleoside phosphorylase revealed a Cupin fold.
Proteins, 90, 2022
|
|
7EYL
 
 | | Crystal structure of Salmonella enterica ppnP | | Descriptor: | Pyrimidine/purine nucleoside phosphorylase | | Authors: | Wen, Y, Wu, B.X. | | Deposit date: | 2021-05-31 | | Release date: | 2022-02-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structures of a new class of pyrimidine/purine nucleoside phosphorylase revealed a Cupin fold.
Proteins, 90, 2022
|
|
7EYM
 
 | | Crystal structure of Vibrio cholerae ppnP | | Descriptor: | Pyrimidine/purine nucleoside phosphorylase | | Authors: | Wen, Y, Wu, B.X. | | Deposit date: | 2021-05-31 | | Release date: | 2022-02-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Crystal structures of a new class of pyrimidine/purine nucleoside phosphorylase revealed a Cupin fold.
Proteins, 90, 2022
|
|
7YV3
 
 | |
