2G4L
 
 | | Anomalous substructure of hydroxynitrile lyase | | Descriptor: | (S)-acetone-cyanohydrin lyase, CHLORIDE ION, SULFATE ION | | Authors: | Mueller-Dieckmann, C, Weiss, M.S. | | Deposit date: | 2006-02-22 | | Release date: | 2007-02-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | On the routine use of soft X-rays in macromolecular crystallography. Part IV. Efficient determination of anomalous substructures in biomacromolecules using longer X-ray wavelengths.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2G4N
 
 | | Anomalous substructure of alpha-lactalbumin | | Descriptor: | Alpha-lactalbumin, CALCIUM ION, POTASSIUM ION | | Authors: | Mueller-Dieckmann, C, Weiss, M.S. | | Deposit date: | 2006-02-22 | | Release date: | 2007-02-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | On the routine use of soft X-rays in macromolecular crystallography. Part IV. Efficient determination of anomalous substructures in biomacromolecules using longer X-ray wavelengths.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2G4Y
 
 | | structure of thaumatin at 2.0 A wavelength | | Descriptor: | D(-)-TARTARIC ACID, Thaumatin-1 | | Authors: | Mueller-Dieckmann, C, Weiss, M.S. | | Deposit date: | 2006-02-22 | | Release date: | 2007-02-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | On the routine use of soft X-rays in macromolecular crystallography. Part IV. Efficient determination of anomalous substructures in biomacromolecules using longer X-ray wavelengths.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2FP0
 
 | | human ADP-ribosylhydrolase 3 | | Descriptor: | ADP-ribosylhydrolase like 2, MAGNESIUM ION | | Authors: | Mueller-Dieckmann, C, Weiss, M.S, Koch-Nolte, F. | | Deposit date: | 2006-01-15 | | Release date: | 2006-10-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The structure of human ADP-ribosylhydrolase 3 (ARH3) provides insights into the reversibility of protein ADP-ribosylation.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2FOZ
 
 | | human ADP-ribosylhydrolase 3 | | Descriptor: | ADP-ribosylhydrolase like 2, MAGNESIUM ION | | Authors: | Mueller-Dieckmann, C, Weiss, M.S, Koch-Nolte, F. | | Deposit date: | 2006-01-15 | | Release date: | 2006-10-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of human ADP-ribosylhydrolase 3 (ARH3) provides insights into the reversibility of protein ADP-ribosylation.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2G4Z
 
 | | anomalous substructure of thermolysin | | Descriptor: | CALCIUM ION, CHLORIDE ION, SULFATE ION, ... | | Authors: | Mueller-Dieckmann, C, Weiss, M.S. | | Deposit date: | 2006-02-22 | | Release date: | 2007-02-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | On the routine use of soft X-rays in macromolecular crystallography. Part IV. Efficient determination of anomalous substructures in biomacromolecules using longer X-ray wavelengths.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
8OGN
 
 | | PanDDA analysis group deposition -- CdaA in complex with fragment F2X-Entry A09 | | Descriptor: | Cyclic di-AMP synthase CdaA, MAGNESIUM ION, [1,2]thiazolo[5,4-b]pyridin-3-amine | | Authors: | Garbers, T.B, Neumann, P, Wollenhaupt, J, Weiss, M.S, Ficner, R. | | Deposit date: | 2023-03-20 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | CdaA in complex with fragment F2X-Entry A09
To Be Published
|
|
8OGM
 
 | |
4HZG
 
 | | Structure of haloalkane dehalogenase DhaA from Rhodococcus rhodochrous | | Descriptor: | CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Stsiapanava, A, Weiss, M.S, Mesters, J.R, Kuta Smatanova, I. | | Deposit date: | 2012-11-15 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystallographic analysis of 1,2,3-trichloropropane biodegradation by the haloalkane dehalogenase DhaA31.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8BM8
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 11 | | Descriptor: | Ephrin type-A receptor 2, ~{N}-(3-cyanophenyl)-4-methyl-3-[(1-methyl-6-pyridin-3-yl-pyrazolo[3,4-d]pyrimidin-4-yl)amino]benzamide | | Authors: | Linhard, V, Witt, K, Gande, S, Wollenhaupt, J, Lennartz, F, Weiss, M.S, Schwalbe, H. | | Deposit date: | 2022-11-10 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | (placeholder, will be filled after publication)
To Be Published
|
|
4A6N
 
 | | STRUCTURE OF THE TETRACYCLINE DEGRADING MONOOXYGENASE TETX IN COMPLEX WITH TIGECYCLINE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, TETX2 PROTEIN, ... | | Authors: | Volkers, G, Palm, G.J, Weiss, M.S, Hinrichs, W. | | Deposit date: | 2011-11-07 | | Release date: | 2012-11-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Putative Dioxygen-Binding Sites and Recognition of Tigecycline and Minocycline in the Tetracycline-Degrading Monooxygenase Tetx
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4A99
 
 | | STRUCTURE OF THE TETRACYCLINE DEGRADING MONOOXYGENASE TETX IN COMPLEX WITH MINOCYCLINE | | Descriptor: | (4S,4AS,5AR,12AS)-4,7-BIS(DIMETHYLAMINO)-3,10,12,12A-TETRAHYDROXY-1,11-DIOXO-1,4,4A,5,5A,6,11,12A-OCTAHYDROTETRACENE-2- CARBOXAMIDE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Volkers, G, Palm, G.J, Weiss, M.S, Hinrichs, W. | | Deposit date: | 2011-11-25 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Putative Dioxygen-Binding Sites and Recognition of Tigecycline and Minocycline in the Tetracycline-Degrading Monooxygenase Tetx
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2W93
 
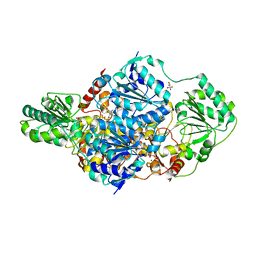 | | Crystal structure of the Saccharomyces cerevisiae pyruvate decarboxylase variant E477Q in complex with the surrogate pyruvamide | | Descriptor: | (1S,2S)-1-amino-1,2-dihydroxypropan-1-olate, MAGNESIUM ION, PYRUVATE DECARBOXYLASE ISOZYME 1, ... | | Authors: | Kutter, S, Weiss, M.S, Konig, S. | | Deposit date: | 2009-01-21 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Allosteric Activation of Pyruvate Decarboxylases. A Never-Ending Story.
J.Mol.Catal., B Enzym., 61, 2014
|
|
4Y36
 
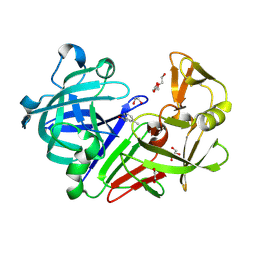 | | Endothiapepsin in complex with fragment 4 | | Descriptor: | 1,2-ETHANEDIOL, 3-methylbenzohydrazide, Endothiapepsin, ... | | Authors: | Radeva, N, Uehlein, M, Weiss, M.S, Heine, A, Klebe, G. | | Deposit date: | 2015-02-10 | | Release date: | 2016-02-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
4Y3P
 
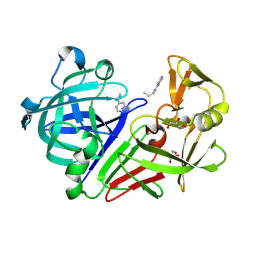 | | Endothiapepsin in complex with fragment 17 | | Descriptor: | 1-(6-methylpyrimidin-4-yl)azepane, Endothiapepsin, GLYCEROL | | Authors: | Radeva, N, Uehlein, M, Weiss, M.S, Heine, A, Klebe, G. | | Deposit date: | 2015-02-10 | | Release date: | 2016-02-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
4Y3E
 
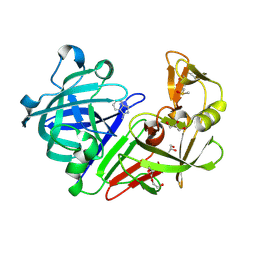 | | Endothiapepsin in complex with fragment 5 | | Descriptor: | 1H-isoindol-3-amine, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Radeva, N, Uehlein, M, Weiss, M.S, Heine, A, Klebe, G. | | Deposit date: | 2015-02-10 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
4Y3W
 
 | | Endothiapepsin in complex with fragment 35 | | Descriptor: | (2R)-2-methyl-2-(morpholin-4-yl)butan-1-amine, 1,2-ETHANEDIOL, Endothiapepsin, ... | | Authors: | Radeva, N, Uehlein, M, Weiss, M.S, Heine, A, Klebe, G. | | Deposit date: | 2015-02-10 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
4Y3Z
 
 | | Endothiapepsin in complex with fragment 41 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Endothiapepsin, ... | | Authors: | Radeva, N, Uehlein, M, Weiss, M.S, Heine, A, Klebe, G. | | Deposit date: | 2015-02-10 | | Release date: | 2016-02-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
4Y43
 
 | | Endothiapepsin in complex with fragment 42 | | Descriptor: | (1E,2E)-N,N'-di(propan-2-yl)cyclohepta-3,5-diene-1,2-diimine, 1,2-ETHANEDIOL, Endothiapepsin, ... | | Authors: | Radeva, N, Uehlein, M, Weiss, M.S, Heine, A, Klebe, G. | | Deposit date: | 2015-02-10 | | Release date: | 2016-02-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
4Y4U
 
 | | Endothiapepsin in complex with fragment 14 | | Descriptor: | 1-(4-bromo-2-chlorophenyl)-3-methylthiourea, DI(HYDROXYETHYL)ETHER, Endothiapepsin, ... | | Authors: | Radeva, N, Uehlein, M, Weiss, M.S, Heine, A, Klebe, G. | | Deposit date: | 2015-02-11 | | Release date: | 2016-03-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.754 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
7FLP
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P05F11 from the F2X-Universal Library | | Descriptor: | 2-methoxybenzamide, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FN6
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P06H06 from the F2X-Universal Library | | Descriptor: | A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8, ethyl (3-methylanilino)(oxo)acetate | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FLT
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P05G04 from the F2X-Universal Library | | Descriptor: | (1S)-1-(3-fluoro-4-methoxyphenyl)ethan-1-amine, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FOR
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P08C12 from the F2X-Universal Library | | Descriptor: | (2S)-1-(2-bromophenoxy)-3-(pyrrolidin-1-yl)propan-2-ol, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FP0
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P08F02 from the F2X-Universal Library | | Descriptor: | 1-cyclopropylimidazolidin-2-one, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
