6NZ1
 
 | |
6O0I
 
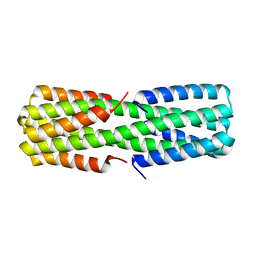
 | | NMR ensemble of computationally designed protein XAA | | Descriptor: | Design construct XAA | | Authors: | Wei, K.Y, Moschidi, D, Nerli, S, Sgourakis, N, Baker, D. | | Deposit date: | 2019-02-16 | | Release date: | 2020-04-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Computational design of closely related proteins that adopt two well-defined but structurally divergent folds.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6NY8
 
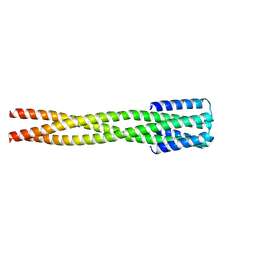 | |
6NYE
 
 | |
6NX2
 
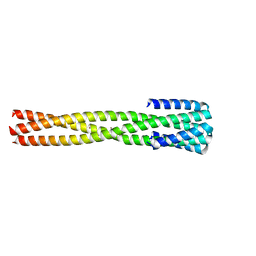 | | Crystal structure of computationally designed protein AAA | | Descriptor: | BROMIDE ION, Design construct AAA | | Authors: | Wei, K.Y, Bick, M.J. | | Deposit date: | 2019-02-07 | | Release date: | 2020-04-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Computational design of closely related proteins that adopt two well-defined but structurally divergent folds.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6NYI
 
 | |
6NYK
 
 | |
6O0C
 
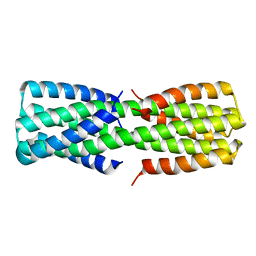 | | NMR ensemble of computationally designed protein XAA_GVDQ mutant M4L | | Descriptor: | Design construct XAA_GVDQ mutant M4L | | Authors: | Wei, K.Y, Moschidi, D, Nerli, S, Sgourakis, N, Baker, D. | | Deposit date: | 2019-02-15 | | Release date: | 2020-04-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Computational design of closely related proteins that adopt two well-defined but structurally divergent folds.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6NXM
 
 | |
6NZ3
 
 | |
3V50
 
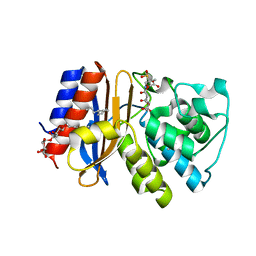 | | Complex of SHV S130G mutant beta-lactamase complexed to SA2-13 | | Descriptor: | (3R)-4-[(4-CARBOXYBUTANOYL)OXY]-N-[(1E)-3-OXOPROP-1-EN-1-YL]-3-SULFINO-D-VALINE, Beta-lactamase, CYCLOHEXYL-HEXYL-BETA-D-MALTOSIDE | | Authors: | Wei, K, van den Akker, F. | | Deposit date: | 2011-12-15 | | Release date: | 2012-08-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The importance of the trans-enamine intermediate as a beta-lactamase inhibition strategy probed in inhibitor-resistant SHV beta-lactamase variants.
Chemmedchem, 7, 2012
|
|
7LVM
 
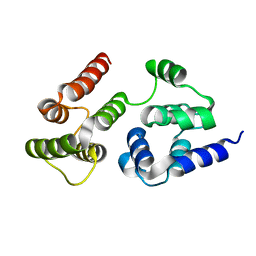 | |
7LVJ
 
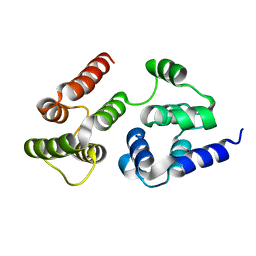 | | CASP8 isoform G DED domain | | Descriptor: | Isoform 9 of Caspase-8 | | Authors: | Weichert, K, Lu, F, Kodandapani, L, Sauder, J.M. | | Deposit date: | 2021-02-25 | | Release date: | 2022-03-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Caspase-8 Variant G Regulates Rheumatoid Arthritis Fibroblast-Like Synoviocyte Aggressive Behavior.
ACR Open Rheumatol, 4, 2022
|
|
1H92
 
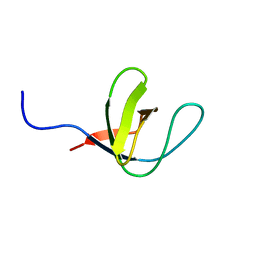 | | SH3 domain of human Lck tyrosine kinase | | Descriptor: | PROTO-ONCOGENE TYROSINE-PROTEIN KINASE LCK | | Authors: | Schweimer, K, Hoffmann, S, Friedrich, U, Biesinger, B, Roesch, P, Sticht, H. | | Deposit date: | 2001-02-22 | | Release date: | 2001-10-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Investigation of the Binding of a Herpesviral Protein to the SH3 Domain of Tyrosine Kinase Lck
Biochemistry, 41, 2002
|
|
1H7V
 
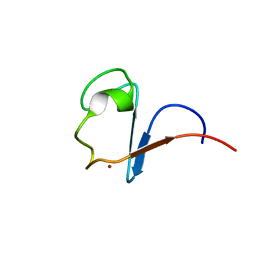 | | Rubredoxin from Guillardia Theta | | Descriptor: | RUBREDOXIN, ZINC ION | | Authors: | Schweimer, K, Hoffmann, S, Wastl, J, Maier, U.G, Roesch, P, Sticht, H. | | Deposit date: | 2001-01-16 | | Release date: | 2002-01-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a zinc substituted eukaryotic rubredoxin from the cryptomonad alga Guillardia theta.
Protein Sci., 9, 2000
|
|
6YEP
 
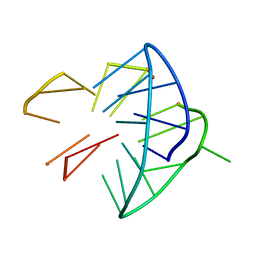 | |
6YCV
 
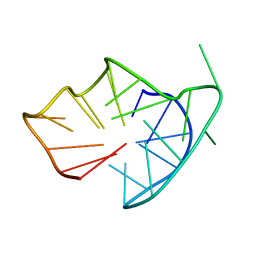 | |
6R3C
 
 | |
3S96
 
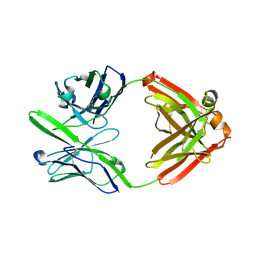 | | Crystal structure of 3B5H10 | | Descriptor: | 3B5H10 FAB heavy chain, 3B5H10 FAB light chain | | Authors: | Weisgraber, K, Peters-Libeu, C, Rutenber, E, Newhouse, Y, Finkbeiner, S. | | Deposit date: | 2011-05-31 | | Release date: | 2012-02-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Disease-associated polyglutamine stretches in monomeric huntingtin adopt a compact structure.
J.Mol.Biol., 421, 2012
|
|
1B6F
 
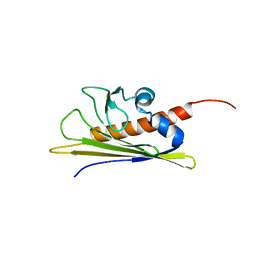 | | BIRCH POLLEN ALLERGEN BET V 1 | | Descriptor: | PROTEIN (MAJOR POLLEN ALLERGEN BET V 1-A) | | Authors: | Schweimer, K, Sticht, H, Boehm, M, Roesch, P. | | Deposit date: | 1999-01-13 | | Release date: | 2000-01-17 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR Spectroscopy Reveals Common Structural Features of the Birch Pollen Allergen Bet v 1 and the cherry allergen Pru a 1
APPL.MAGN.RESON., 17, 1999
|
|
2BZ2
 
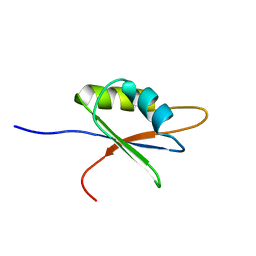 | | Solution structure of NELF E RRM | | Descriptor: | NEGATIVE ELONGATION FACTOR E | | Authors: | Schweimer, K, Rao, J.N, Neumann, L, Rosch, P, Wohrl, B.M. | | Deposit date: | 2005-08-10 | | Release date: | 2006-08-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural studies on the RNA-recognition motif of NELF E, a cellular negative transcription elongation factor involved in the regulation of HIV transcription.
Biochem. J., 400, 2006
|
|
1DX8
 
 | | Rubredoxin from Guillardia theta | | Descriptor: | RUBREDOXIN, ZINC ION | | Authors: | Schweimer, K, Hoffmann, S, Wastl, J, Maier, U.G, Roesch, P, Sticht, H. | | Deposit date: | 1999-12-23 | | Release date: | 2000-01-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a zinc substituted eukaryotic rubredoxin from the cryptomonad alga Guillardia theta.
Protein Sci., 9, 2000
|
|
1E0Z
 
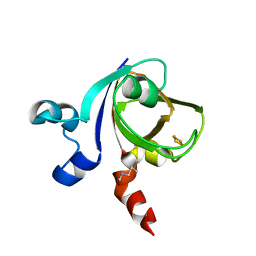 | | [2Fe-2S]-Ferredoxin from Halobacterium salinarum | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN | | Authors: | Schweimer, K, Marg, B, Oesterhelt, D, Roesch, P, Sticht, H. | | Deposit date: | 2000-04-11 | | Release date: | 2001-04-12 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | A Two-Alpha-Helix Extra Domain Mediates the Halophilic Character of a Plant-Type Ferredoxin from Halophilic Archaea.
Biochemistry, 44, 2005
|
|
2JVV
 
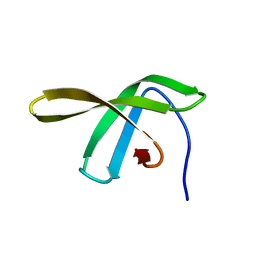 | |
2K06
 
 | |
