6FVX
 
 | | 26S proteasome, s5 state | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome regulatory subunit 4 homolog, 26S proteasome regulatory subunit 6A, ... | | Authors: | Eisele, M.R, Reed, R.G, Rudack, T, Schweitzer, A, Beck, F, Nagy, I, Pfeifer, G, Plitzko, J.M, Baumeister, W, Tomko, R.J, Sakata, E. | | Deposit date: | 2018-03-05 | | Release date: | 2018-08-22 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Expanded Coverage of the 26S Proteasome Conformational Landscape Reveals Mechanisms of Peptidase Gating.
Cell Rep, 24, 2018
|
|
7S4L
 
 | | CryoEM structure of Methylotuvimicrobium alcaliphilum 20Z pMMO in a POPC nanodisc at 2.46 Angstrom resolution | | Descriptor: | (S)-2,3-bis(hexanoyloxy)propyl(2-(trimethylammonio)ethyl)phosphate, 1,2-dihexanoyl-sn-glycero-3-phosphocholine, COPPER (II) ION, ... | | Authors: | Koo, C.W, Rosenzweig, A.C. | | Deposit date: | 2021-09-09 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | Recovery of particulate methane monooxygenase structure and activity in a lipid bilayer.
Science, 375, 2022
|
|
7S4I
 
 | | CryoEM structure of Methylococcus capsulatus (Bath) pMMO in a native lipid nanodisc at 2.26 Angstrom resolution | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dihexanoyl-sn-glycero-3-phosphocholine, Ammonia monooxygenase/methane monooxygenase, ... | | Authors: | Koo, C.W, Rosenzweig, A.C. | | Deposit date: | 2021-09-09 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.26 Å) | | Cite: | Recovery of particulate methane monooxygenase structure and activity in a lipid bilayer.
Science, 375, 2022
|
|
6FVT
 
 | | 26S proteasome, s1 state | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome regulatory subunit 4 homolog, 26S proteasome regulatory subunit 6A, ... | | Authors: | Eisele, M.R, Reed, R.G, Rudack, T, Schweitzer, A, Beck, F, Nagy, I, Pfeifer, G, Plitzko, J.M, Baumeister, W, Tomko, R.J, Sakata, E. | | Deposit date: | 2018-03-05 | | Release date: | 2018-08-22 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Expanded Coverage of the 26S Proteasome Conformational Landscape Reveals Mechanisms of Peptidase Gating.
Cell Rep, 24, 2018
|
|
7S4J
 
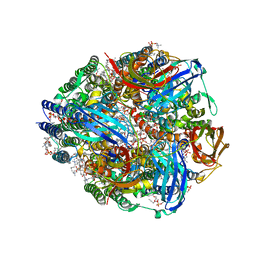 | | CryoEM structure of Methylococcus capsulatus (Bath) pMMO in a native lipid nanodisc at 2.16 Angstrom resolution | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dihexanoyl-sn-glycero-3-phosphocholine, Ammonia monooxygenase/methane monooxygenase, ... | | Authors: | Koo, C.W, Rosenzweig, A.C. | | Deposit date: | 2021-09-09 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.16 Å) | | Cite: | Recovery of particulate methane monooxygenase structure and activity in a lipid bilayer.
Science, 375, 2022
|
|
7S4K
 
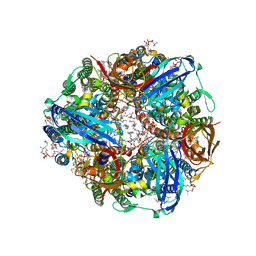 | | CryoEM structure of Methylococcus capsulatus (Bath) pMMO in a native lipid nanodisc at 2.34 Angstrom resolution | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dihexanoyl-sn-glycero-3-phosphocholine, Ammonia monooxygenase/methane monooxygenase, ... | | Authors: | Koo, C.W, Rosenzweig, A.C. | | Deposit date: | 2021-09-09 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | Recovery of particulate methane monooxygenase structure and activity in a lipid bilayer.
Science, 375, 2022
|
|
7S4H
 
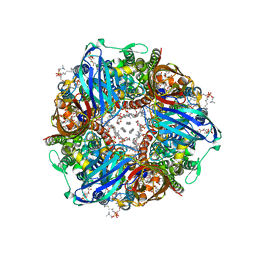 | | CryoEM structure of Methylococcus capsulatus (Bath) pMMO in a native lipid nanodisc at 2.14 Angstrom resolution | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dihexanoyl-sn-glycero-3-phosphocholine, Ammonia monooxygenase/methane monooxygenase, ... | | Authors: | Koo, C.W, Rosenzweig, A.C. | | Deposit date: | 2021-09-08 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.14 Å) | | Cite: | Recovery of particulate methane monooxygenase structure and activity in a lipid bilayer.
Science, 375, 2022
|
|
7S4M
 
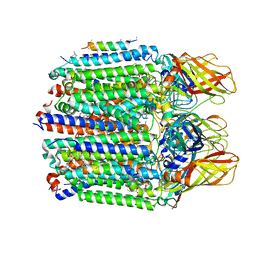 | |
6FVY
 
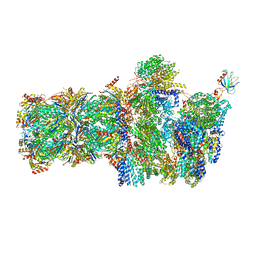 | | 26S proteasome, s6 state | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome regulatory subunit 4 homolog, 26S proteasome regulatory subunit 6A, ... | | Authors: | Eisele, M.R, Reed, R.G, Rudack, T, Schweitzer, A, Beck, F, Nagy, I, Pfeifer, G, Plitzko, J.M, Baumeister, W, Tomko, R.J, Sakata, E. | | Deposit date: | 2018-03-05 | | Release date: | 2018-08-22 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Expanded Coverage of the 26S Proteasome Conformational Landscape Reveals Mechanisms of Peptidase Gating.
Cell Rep, 24, 2018
|
|
3RGB
 
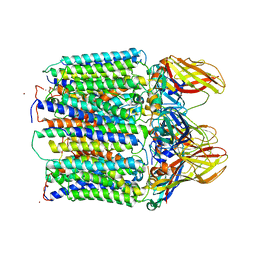 | |
6FVW
 
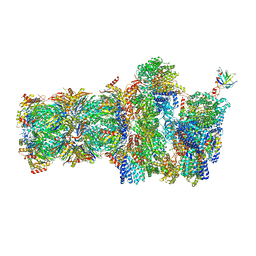 | | 26S proteasome, s4 state | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome regulatory subunit 4 homolog, 26S proteasome regulatory subunit 6A, ... | | Authors: | Eisele, M.R, Reed, R.G, Rudack, T, Schweitzer, A, Beck, F, Nagy, I, Pfeifer, G, Plitzko, J.M, Baumeister, W, Tomko, R.J, Sakata, E. | | Deposit date: | 2018-03-05 | | Release date: | 2018-08-29 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Expanded Coverage of the 26S Proteasome Conformational Landscape Reveals Mechanisms of Peptidase Gating.
Cell Rep, 24, 2018
|
|
4L1Y
 
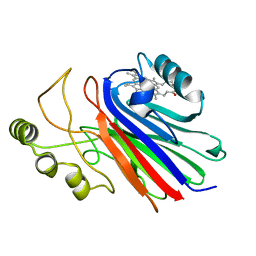 | |
7T4P
 
 | | CryoEM structure of Methylococcus capsulatus (Bath) pMMO treated with potassium cyanide and copper in a native lipid nanodisc at 3.62 Angstrom resolution | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dihexanoyl-sn-glycero-3-phosphocholine, Ammonia monooxygenase/methane monooxygenase, ... | | Authors: | Koo, C.W, Rosenzweig, A.C. | | Deposit date: | 2021-12-10 | | Release date: | 2022-03-30 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Recovery of particulate methane monooxygenase structure and activity in a lipid bilayer.
Science, 375, 2022
|
|
7T4O
 
 | |
3SKX
 
 | | Crystal structure of the ATP binding domain of Archaeoglobus fulgidus COPB | | Descriptor: | ACETATE ION, Copper-exporting P-type ATPase B | | Authors: | Jayakanthan, S, Roberts, S.A, Weichsel, A, Arguello, J.M, McEvoy, M.M. | | Deposit date: | 2011-06-23 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Conformations of the apo-, substrate-bound and phosphate-bound ATP-binding domain of the Cu(II) ATPase CopB illustrate coupling of domain movement to the catalytic cycle.
Biosci.Rep., 32, 2012
|
|
6RI7
 
 | | Cryo-EM structure of E. coli RNA polymerase elongation complex bound to GreB transcription factor | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Abdelkareem, M, Saint-Andre, C, Takacs, M, Papai, G, Crucifix, C, Guo, X, Ortiz, J, Weixlbaumer, A. | | Deposit date: | 2019-04-23 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Basis of Transcription: RNA Polymerase Backtracking and Its Reactivation.
Mol.Cell, 75, 2019
|
|
6RH3
 
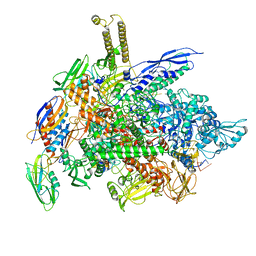 | | Cryo-EM structure of E. coli RNA polymerase elongation complex bound to CTP substrate | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Abdelkareem, M, Saint-Andre, C, Takacs, M, Papai, G, Crucifix, C, Guo, X, Ortiz, J, Weixlbaumer, A. | | Deposit date: | 2019-04-18 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Basis of Transcription: RNA Polymerase Backtracking and Its Reactivation.
Mol.Cell, 75, 2019
|
|
5ICQ
 
 | |
6RI9
 
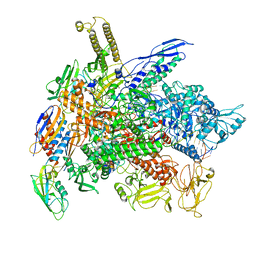 | | Cryo-EM structure of E. coli RNA polymerase backtracked elongation complex in non-swiveled state | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Abdelkareem, M, Saint-Andre, C, Takacs, M, Papai, G, Crucifix, C, Guo, X, Ortiz, J, Weixlbaumer, A. | | Deposit date: | 2019-04-23 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural Basis of Transcription: RNA Polymerase Backtracking and Its Reactivation.
Mol.Cell, 75, 2019
|
|
6RIP
 
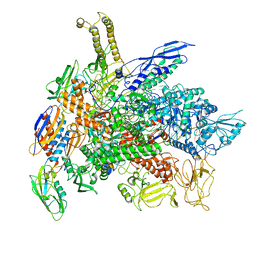 | | Cryo-EM structure of E. coli RNA polymerase backtracked elongation complex in swiveled state | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Abdelkareem, M, Saint-Andre, C, Takacs, M, Papai, G, Crucifix, C, Guo, X, Ortiz, J, Weixlbaumer, A. | | Deposit date: | 2019-04-24 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural Basis of Transcription: RNA Polymerase Backtracking and Its Reactivation.
Mol.Cell, 75, 2019
|
|
6FVV
 
 | | 26S proteasome, s3 state | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome regulatory subunit 4 homolog, 26S proteasome regulatory subunit 6A, ... | | Authors: | Eisele, M.R, Reed, R.G, Rudack, T, Schweitzer, A, Beck, F, Nagy, I, Pfeifer, G, Plitzko, J.M, Baumeister, W, Tomko, R.J, Sakata, E. | | Deposit date: | 2018-03-05 | | Release date: | 2018-08-22 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Expanded Coverage of the 26S Proteasome Conformational Landscape Reveals Mechanisms of Peptidase Gating.
Cell Rep, 24, 2018
|
|
7TCU
 
 | | Methanobactin biosynthetic protein complex of MbnB and MbnC from Methylosinus trichosporium OB3b at 2.31 Angstrom resolution | | Descriptor: | FE (III) ION, Methanobactin biosynthesis cassette protein MbnB, Methanobactin biosynthesis cassette protein MbnC, ... | | Authors: | Park, Y, Reyes, R.M, Rosenzweig, A.C. | | Deposit date: | 2021-12-28 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | A mixed-valent Fe(II)Fe(III) species converts cysteine to an oxazolone/thioamide pair in methanobactin biosynthesis.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7TCW
 
 | | Methanobactin biosynthetic protein complex of MbnB and MbnC from Methylosinus trichosporium OB3b, H210S mutant | | Descriptor: | FE (III) ION, Methanobactin biosynthesis cassette protein MbnB, Methanobactin biosynthesis cassette protein MbnC, ... | | Authors: | Park, Y, Reyes, R.M, Rosenzweig, A.C. | | Deposit date: | 2021-12-28 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | A mixed-valent Fe(II)Fe(III) species converts cysteine to an oxazolone/thioamide pair in methanobactin biosynthesis.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7TCX
 
 | | Methanobactin biosynthetic protein complex of MbnB and MbnC from Methylosinus trichosporium OB3b at 2.21 Angstrom resolution | | Descriptor: | FE (III) ION, Methanobactin biosynthesis cassette protein MbnB, Methanobactin biosynthesis cassette protein MbnC, ... | | Authors: | Park, Y, Reyes, R.M, Rosenzweig, A.C. | | Deposit date: | 2021-12-28 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | A mixed-valent Fe(II)Fe(III) species converts cysteine to an oxazolone/thioamide pair in methanobactin biosynthesis.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
3N37
 
 | | Ribonucleotide Reductase Dimanganese(II)-NrdF from Escherichia coli | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Ribonucleoside-diphosphate reductase 2 subunit beta | | Authors: | Boal, A.K, Cotruvo Jr, J.A, Stubbe, J, Rosenzweig, A.C. | | Deposit date: | 2010-05-19 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for activation of class Ib ribonucleotide reductase.
Science, 329, 2010
|
|
