7PY3
 
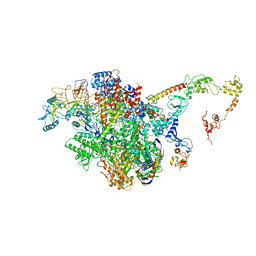 | | CryoEM structure of E.coli RNA polymerase elongation complex bound to NusA (the consensus NusA-EC) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zhu, C, Guo, X, Weixlbaumer, A. | | Deposit date: | 2021-10-08 | | Release date: | 2022-03-23 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Transcription factors modulate RNA polymerase conformational equilibrium.
Nat Commun, 13, 2022
|
|
7PYJ
 
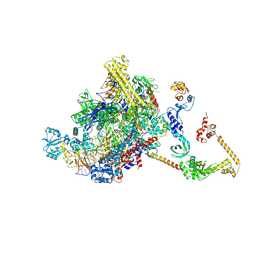 | | CryoEM structure of E.coli RNA polymerase elongation complex bound to NusA (NusA elongation complex in less-swiveled conformation) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zhu, C, Guo, X, Weixlbaumer, A. | | Deposit date: | 2021-10-10 | | Release date: | 2022-03-23 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Transcription factors modulate RNA polymerase conformational equilibrium.
Nat Commun, 13, 2022
|
|
7PY6
 
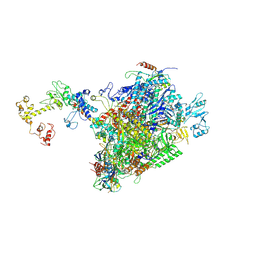 | | CryoEM structure of E.coli RNA polymerase elongation complex bound to NusA and NusG (NusA and NusG elongation complex in less-swiveled conformation) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zhu, C, Guo, X, Weixlbaumer, A. | | Deposit date: | 2021-10-09 | | Release date: | 2022-03-23 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Transcription factors modulate RNA polymerase conformational equilibrium.
Nat Commun, 13, 2022
|
|
9CL5
 
 | | particulate methane monooxygenase in native membranes | | Descriptor: | COPPER (II) ION, Methane monooxygenase/ammonia monooxygenase subunit A, Methane monooxygenase/ammonia monooxygenase subunit B, ... | | Authors: | Tucci, F.J, Rosenzweig, A.C. | | Deposit date: | 2024-07-10 | | Release date: | 2025-01-08 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Structures of methane and ammonia monooxygenases in native membranes.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8SR5
 
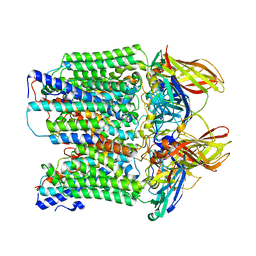 | |
2LVN
 
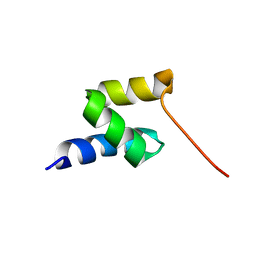 | | Structure of the gp78 CUE domain | | Descriptor: | E3 ubiquitin-protein ligase AMFR | | Authors: | Liu, S, Chen, Y, Huang, T, Tarasov, S.G, King, A, Li, J, Weissman, A.M, Byrd, R.A, Das, R. | | Deposit date: | 2012-07-09 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Promiscuous Interactions of gp78 E3 Ligase CUE Domain with Polyubiquitin Chains.
Structure, 20, 2012
|
|
2LVO
 
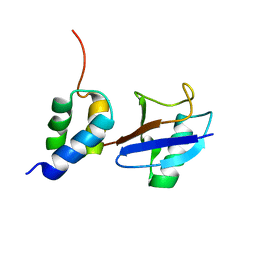 | | Structure of the gp78CUE domain bound to monubiquitin | | Descriptor: | E3 ubiquitin-protein ligase AMFR, Ubiquitin | | Authors: | Liu, S, Chen, Y, Huang, T, Tarasov, S.G, King, A, Li, J, Weissman, A.M, Byrd, R.A, Das, R. | | Deposit date: | 2012-07-09 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Promiscuous Interactions of gp78 E3 Ligase CUE Domain with Polyubiquitin Chains.
Structure, 20, 2012
|
|
2LXH
 
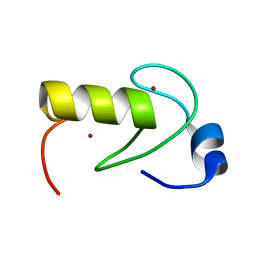 | | NMR structure of the RING domain in ubiquitin ligase gp78 | | Descriptor: | E3 ubiquitin-protein ligase AMFR, ZINC ION | | Authors: | Das, R, Linag, Y, Mariano, J, Li, J, Huang, T, King, A, Weissman, A, Ji, X, Byrd, R. | | Deposit date: | 2012-08-27 | | Release date: | 2013-08-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Allosteric regulation of E2:E3 interactions promote a processive ubiquitination machine.
Embo J., 32, 2013
|
|
2LVP
 
 | | gp78CUE domain bound to the distal ubiquitin of K48-linked diubiquitin | | Descriptor: | E3 ubiquitin-protein ligase AMFR, Ubiquitin | | Authors: | Liu, S, Chen, Y, Huang, T, Tarasov, S.G, King, A, Li, J, Weissman, A.M, Byrd, R.A, Das, R. | | Deposit date: | 2012-07-09 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Promiscuous Interactions of gp78 E3 Ligase CUE Domain with Polyubiquitin Chains.
Structure, 20, 2012
|
|
2LVQ
 
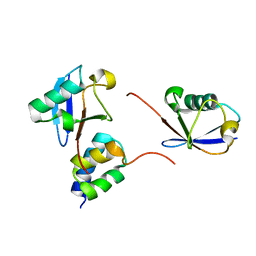 | | gp78CUE domain bound to the proximal ubiquitin of K48-linked diubiquitin | | Descriptor: | E3 ubiquitin-protein ligase AMFR, Ubiquitin | | Authors: | Liu, S, Chen, Y, Huang, T, Tarasov, S.G, King, A, Li, J, Weissman, A.M, Byrd, R.A, Das, R. | | Deposit date: | 2012-07-09 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Promiscuous Interactions of gp78 E3 Ligase CUE Domain with Polyubiquitin Chains.
Structure, 20, 2012
|
|
2LXP
 
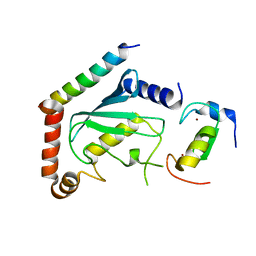 | | NMR structure of two domains in ubiquitin ligase gp78, RING and G2BR, bound to its conjugating enzyme Ube2g | | Descriptor: | E3 ubiquitin-protein ligase AMFR, Ubiquitin-conjugating enzyme E2 G2, ZINC ION | | Authors: | Das, R, Linag, Y, Mariano, J, Li, J, Huang, T, King, A, Weissman, A, Ji, X, Byrd, R. | | Deposit date: | 2012-08-30 | | Release date: | 2013-08-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Allosteric regulation of E2:E3 interactions promote a processive ubiquitination machine.
Embo J., 32, 2013
|
|
7N99
 
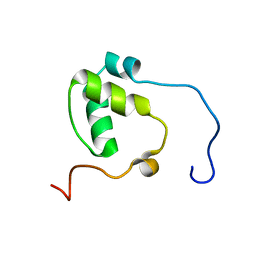 | | SDE2 SAP domain apo structure | | Descriptor: | Isoform 2 of Replication stress response regulator SDE2 | | Authors: | Paung, Y, Weinheimer, A.S, Rageul, J, Khan, A, Ho, B, Tong, M, Alphonse, S, Seeliger, M.A, Kim, H. | | Deposit date: | 2021-06-17 | | Release date: | 2022-10-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Extended DNA-binding interfaces beyond the canonical SAP domain contribute to the function of replication stress regulator SDE2 at DNA replication forks.
J.Biol.Chem., 298, 2022
|
|
2NRM
 
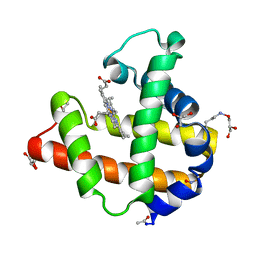 | | S-nitrosylated blackfin tuna myoglobin | | Descriptor: | GLYCEROL, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Schreiter, E.R, Rodr guez, M.M, Weichsel, A, Montfort, W.R, Bonaventura, J. | | Deposit date: | 2006-11-02 | | Release date: | 2007-05-08 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | S-nitrosylation-induced conformational change in blackfin tuna myoglobin.
J.Biol.Chem., 282, 2007
|
|
9CL4
 
 | | Particulate methane monooxygenase in crosslinked, washed native membranes | | Descriptor: | (2R)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(hexadecanoyloxy)propyl (9Z)-heptadec-9-enoate, COPPER (II) ION, Particulate methane monooxygenase alpha subunit, ... | | Authors: | Tucci, F.J, Rosenzweig, A.C. | | Deposit date: | 2024-07-10 | | Release date: | 2025-01-08 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Structures of methane and ammonia monooxygenases in native membranes.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9CL2
 
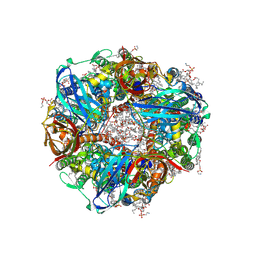 | | Particulate methane monooxygenase in washed native membranes | | Descriptor: | (2R)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(hexadecanoyloxy)propyl (9Z)-heptadec-9-enoate, COPPER (II) ION, Particulate methane monooxygenase alpha subunit, ... | | Authors: | Tucci, F.J, Rosenzweig, A.C. | | Deposit date: | 2024-07-10 | | Release date: | 2025-01-08 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (2.42 Å) | | Cite: | Structures of methane and ammonia monooxygenases in native membranes.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9CL3
 
 | | Particulate methane monooxygenase in unwashed native membranes | | Descriptor: | (2R)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(hexadecanoyloxy)propyl (9Z)-heptadec-9-enoate, COPPER (II) ION, Particulate methane monooxygenase alpha subunit, ... | | Authors: | Tucci, F.J, Rosenzweig, A.C. | | Deposit date: | 2024-07-10 | | Release date: | 2025-01-08 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Structures of methane and ammonia monooxygenases in native membranes.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
2OZO
 
 | | Autoinhibited intact human ZAP-70 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Tyrosine-protein kinase ZAP-70 | | Authors: | Deindl, S, Kadlecek, T.A, Brdicka, T, Cao, X, Weiss, A, Kuriyan, J. | | Deposit date: | 2007-02-26 | | Release date: | 2007-05-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for the Inhibition of Tyrosine Kinase Activity of ZAP-70.
Cell(Cambridge,Mass.), 129, 2007
|
|
2NX0
 
 | | Ferrous nitrosyl blackfin tuna myoglobin | | Descriptor: | Myoglobin, NITRIC OXIDE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Schreiter, E.R, Rodriguez, M.M, Weichsel, A, Montfort, W.R, Bonaventura, J. | | Deposit date: | 2006-11-16 | | Release date: | 2007-05-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | S-nitrosylation-induced conformational change in blackfin tuna myoglobin.
J.Biol.Chem., 282, 2007
|
|
2IN4
 
 | | Crystal Structure of Myoglobin with Charge Neutralized Heme, ZnDMb-dme | | Descriptor: | Myoglobin, SULFATE ION, ZINC(II)-DEUTEROPORPHYRIN DIMETHYLESTER | | Authors: | Wheeler, K.E, Nocek, J.M, Cull, D.A, Yatsunyk, L.A, Rosenzweig, A.C, Hoffman, B.M. | | Deposit date: | 2006-10-05 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Dynamic docking of cytochrome b5 with myoglobin and alpha-hemoglobin: heme-neutralization "squares" and the binding of electron-transfer-reactive configurations.
J.Am.Chem.Soc., 129, 2007
|
|
2NRL
 
 | | Blackfin tuna myoglobin | | Descriptor: | 1,2-ETHANEDIOL, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Schreiter, E.R, Rodr guez, M.M, Weichsel, A, Montfort, W.R, Bonaventura, J. | | Deposit date: | 2006-11-02 | | Release date: | 2007-05-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (0.91 Å) | | Cite: | S-nitrosylation-induced conformational change in blackfin tuna myoglobin.
J.Biol.Chem., 282, 2007
|
|
8OYI
 
 | | particulate methane monooxygenase with 2,2,2-trifluoroethanol bound | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dihexanoyl-sn-glycero-3-phosphocholine, Ammonia monooxygenase/methane monooxygenase, ... | | Authors: | Tucci, F.J, Rosenzweig, A.C. | | Deposit date: | 2023-05-04 | | Release date: | 2023-11-08 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (2.19 Å) | | Cite: | Product analog binding identifies the copper active site of particulate methane monooxygenase.
Nat Catal, 6, 2023
|
|
2H2M
 
 | |
2ROP
 
 | | Solution structure of domains 3 and 4 of human ATP7B | | Descriptor: | Copper-transporting ATPase 2 | | Authors: | Banci, L, Bertini, I, Cantini, F, Rosenzweig, A.C, Yatsunyk, L.A. | | Deposit date: | 2008-04-04 | | Release date: | 2008-10-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Metal binding domains 3 and 4 of the Wilson disease protein: solution structure and interaction with the copper(I) chaperone HAH1
Biochemistry, 47, 2008
|
|
6EWU
 
 | | Solution Structure of Rhabdopeptide NRPS Docking Domain Kj12C-NDD | | Descriptor: | NRPS Kj12C-NDD | | Authors: | Hacker, C, Cai, X, Kegler, C, Zhao, L, Weickhmann, A.K, Bode, H.B, Woehnert, J. | | Deposit date: | 2017-11-06 | | Release date: | 2018-10-31 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | Structure-based redesign of docking domain interactions modulates the product spectrum of a rhabdopeptide-synthesizing NRPS.
Nat Commun, 9, 2018
|
|
6FVU
 
 | | 26S proteasome, s2 state | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome regulatory subunit 4 homolog, 26S proteasome regulatory subunit 6A, ... | | Authors: | Eisele, M.R, Reed, R.G, Rudack, T, Schweitzer, A, Beck, F, Nagy, I, Pfeifer, G, Plitzko, J.M, Baumeister, W, Tomko, R.J, Sakata, E. | | Deposit date: | 2018-03-05 | | Release date: | 2018-08-22 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Expanded Coverage of the 26S Proteasome Conformational Landscape Reveals Mechanisms of Peptidase Gating.
Cell Rep, 24, 2018
|
|
