1E0J
 
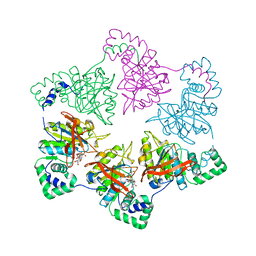 | | gp4d helicase from phage T7 ADPNP complex | | Descriptor: | DNA HELICASE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Singleton, M.R, Sawaya, M.R, Ellenberger, T, Wigley, D.B. | | Deposit date: | 2000-03-30 | | Release date: | 2000-06-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of T7 Gene 4 Ring Helicase Indicates a Mechanism for Sequential Hydrolysis of Nucleotides
Cell(Cambridge,Mass.), 101, 2000
|
|
1K26
 
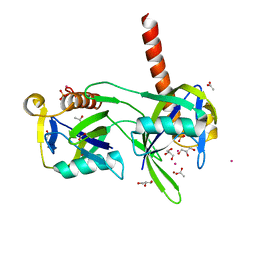 | | Structure of a Nudix Protein from Pyrobaculum aerophilum Solved by the Single Wavelength Anomolous Scattering Method | | Descriptor: | ACETIC ACID, GLYCEROL, IRIDIUM (III) ION, ... | | Authors: | Wang, S, Mura, C, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2001-09-26 | | Release date: | 2002-04-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of a Nudix protein from Pyrobaculum aerophilum reveals a dimer with two intersubunit beta-sheets.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
3Q9G
 
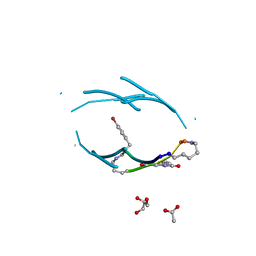 | | VQIVY segment from Alzheimer's tau displayed on 42-membered macrocycle scaffold | | Descriptor: | ACETIC ACID, Cyclic pseudo-peptide VQIV(4BF)(ORN)(HAO)KL(ORN), GLYCEROL | | Authors: | Liu, C, Sawaya, M.R, Eisenberg, D, Nowick, J.S, Cheng, P, Zheng, J. | | Deposit date: | 2011-01-07 | | Release date: | 2011-06-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Characteristics of Amyloid-Related Oligomers Revealed by Crystal Structures of Macrocyclic beta-Sheet Mimics.
J.Am.Chem.Soc., 133, 2011
|
|
1JG4
 
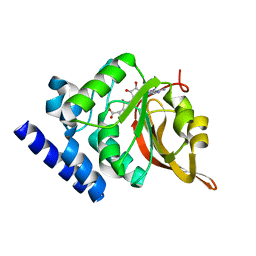 | | Crystal Structure of L-isoaspartyl (D-aspartyl) O-methyltransferase with S-adenosylmethionine | | Descriptor: | S-ADENOSYLMETHIONINE, protein-L-isoaspartate O-methyltransferase | | Authors: | Griffith, S.C, Sawaya, M.R, Boutz, D, Thapar, N, Katz, J, Clarke, S, Yeates, T.O. | | Deposit date: | 2001-06-22 | | Release date: | 2001-11-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of a protein repair methyltransferase from Pyrococcus furiosus with its L-isoaspartyl peptide substrate.
J.Mol.Biol., 313, 2001
|
|
1I8F
 
 | | THE CRYSTAL STRUCTURE OF A HEPTAMERIC ARCHAEAL SM PROTEIN: IMPLICATIONS FOR THE EUKARYOTIC SNRNP CORE | | Descriptor: | GLYCEROL, PUTATIVE SNRNP SM-LIKE PROTEIN | | Authors: | Mura, C, Cascio, D, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2001-03-14 | | Release date: | 2001-05-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The crystal structure of a heptameric archaeal Sm protein: Implications for the eukaryotic snRNP core.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
4OLO
 
 | | Ligand-free structure of the GrpU microcompartment shell protein from Clostridiales bacterium 1_7_47FAA | | Descriptor: | BMC domain protein | | Authors: | Thompson, M.C, Ahmed, H, McCarty, K.N, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2014-01-24 | | Release date: | 2014-07-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of a unique fe-s cluster binding site in a glycyl-radical type microcompartment shell protein.
J.Mol.Biol., 426, 2014
|
|
1K2E
 
 | | crystal structure of a nudix protein from Pyrobaculum aerophilum | | Descriptor: | ACETIC ACID, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Wang, S, Mura, C, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2001-09-26 | | Release date: | 2002-04-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a Nudix protein from Pyrobaculum aerophilum reveals a dimer with two intersubunit beta-sheets.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1JG3
 
 | | Crystal Structure of L-isoaspartyl (D-aspartyl) O-methyltransferase with adenosine & VYP(ISP)HA substrate | | Descriptor: | ADENOSINE, CHLORIDE ION, SODIUM ION, ... | | Authors: | Griffith, S.C, Sawaya, M.R, Boutz, D, Thapar, N, Katz, J, Clarke, S, Yeates, T.O. | | Deposit date: | 2001-06-22 | | Release date: | 2001-11-16 | | Last modified: | 2011-07-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a protein repair methyltransferase from Pyrococcus furiosus with its L-isoaspartyl peptide substrate.
J.Mol.Biol., 313, 2001
|
|
1JG2
 
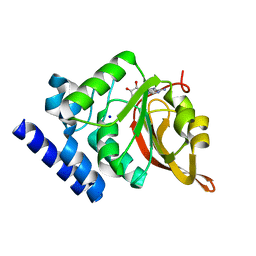 | | Crystal Structure of L-isoaspartyl (D-aspartyl) O-methyltransferase with adenosine | | Descriptor: | ADENOSINE, SODIUM ION, protein-L-isoaspartate O-methyltransferase | | Authors: | Griffith, S.C, Sawaya, M.R, Boutz, D, Thapar, N, Katz, J, Clarke, S, Yeates, T.O. | | Deposit date: | 2001-06-22 | | Release date: | 2001-11-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of a protein repair methyltransferase from Pyrococcus furiosus with its L-isoaspartyl peptide substrate.
J.Mol.Biol., 313, 2001
|
|
1MZ4
 
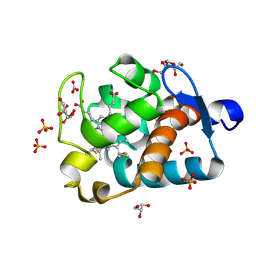 | | Crystal Structure of Cytochrome c550 from Thermosynechococcus elongatus | | Descriptor: | BICARBONATE ION, GLYCEROL, HEME C, ... | | Authors: | Kerfeld, C.A, Sawaya, M.R, Bottin, H, Tran, K.T, Sugiura, M, Kirilovsky, D, Krogmann, D, Yeates, T.O, Boussac, A. | | Deposit date: | 2002-10-05 | | Release date: | 2003-09-23 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and EPR characterization of the soluble form of cytochrome c-550 and of the psbV2 gene product from the cyanobacterium Thermosynechococcus elongatus.
Plant Cell.Physiol., 44, 2003
|
|
4RZT
 
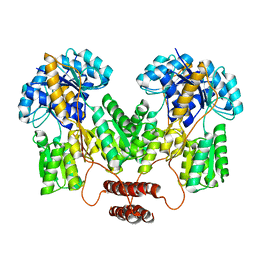 | | Lac repressor engineered to bind sucralose, sucralose-bound tetramer | | Descriptor: | 4-chloro-4-deoxy-alpha-D-galactopyranose-(1-2)-1,6-dichloro-1,6-dideoxy-beta-D-fructofuranose, Lac repressor | | Authors: | Arbing, M.A, Cascio, D, Sawaya, M.R, Kosuri, S, Church, G.M. | | Deposit date: | 2014-12-24 | | Release date: | 2015-12-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Engineering an allosteric transcription factor to respond to new ligands.
Nat.Methods, 13, 2016
|
|
4RO5
 
 | | Crystal structure of the SAT domain from the non-reducing fungal polyketide synthase CazM | | Descriptor: | GLYCEROL, SAT domain from CazM | | Authors: | Winter, J.M, Cascio, D, Sawaya, M.R, Tang, Y. | | Deposit date: | 2014-10-27 | | Release date: | 2015-09-09 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Biochemical and Structural Basis for Controlling Chemical Modularity in Fungal Polyketide Biosynthesis.
J.Am.Chem.Soc., 137, 2015
|
|
4OIJ
 
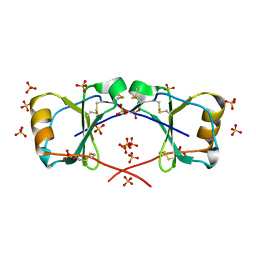 | | X-ray crystal structure of racemic non-glycosylated chemokine Ser-CCL1 | | Descriptor: | C-C motif chemokine 1, D-Ser-CCL1, SULFATE ION | | Authors: | Okamoto, R, Mandal, K, Sawaya, M.R, Kajihara, Y, Yeates, T.O, Kent, S.B.H. | | Deposit date: | 2014-01-19 | | Release date: | 2014-05-07 | | Last modified: | 2014-05-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | (Quasi-)Racemic X-ray Structures of Glycosylated and Non-Glycosylated Forms of the Chemokine Ser-CCL1 Prepared by Total Chemical Synthesis.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4OLR
 
 | | [Leu-5]-Enkephalin mutant - YVVFV | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, [Leu-5]-Enkephalin mutant - YVVFV | | Authors: | Sangwan, S, Eisenberg, D, Sawaya, M.R, Do, T.D, Bowers, M.T, Lapointe, N.E, Teplow, D.B, Feinstein, S.C. | | Deposit date: | 2014-01-24 | | Release date: | 2014-07-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Factors that drive Peptide assembly from native to amyloid structures: experimental and theoretical analysis of [leu-5]-enkephalin mutants.
J.Phys.Chem.B, 118, 2014
|
|
4P2S
 
 | | Alanine Scanning Mutagenesis Identifies an Asparagine-Arginine-Lysine Triad Essential to Assembly of the Shell of the Pdu Microcompartment | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Putative propanediol utilization protein PduA, ... | | Authors: | Sinha, S, Cheng, S, Sung, Y.W, McNamara, D.E, Sawaya, M.R, Yeates, T.O, Bobik, T.A. | | Deposit date: | 2014-03-03 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Alanine Scanning Mutagenesis Identifies an Asparagine-Arginine-Lysine Triad Essential to Assembly of the Shell of the Pdu Microcompartment.
J.Mol.Biol., 426, 2014
|
|
4OIK
 
 | | (Quasi-)Racemic X-ray crystal structure of glycosylated chemokine Ser-CCL1. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C-C motif chemokine 1, CITRIC ACID, ... | | Authors: | Okamoto, R, Mandal, K, Sawaya, M.R, Kajihara, Y, Yeates, T.O, Kent, S.B.H. | | Deposit date: | 2014-01-19 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | (Quasi-)Racemic X-ray Structures of Glycosylated and Non-Glycosylated Forms of the Chemokine Ser-CCL1 Prepared by Total Chemical Synthesis.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
1S3Q
 
 | | Crystal structures of a novel open pore ferritin from the hyperthermophilic Archaeon Archaeoglobus fulgidus | | Descriptor: | ZINC ION, ferritin | | Authors: | Johnson, E, Cascio, D, Sawaya, M, Schroeder, I. | | Deposit date: | 2004-01-13 | | Release date: | 2005-04-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of a tetrahedral open pore ferritin from the hyperthermophilic archaeon Archaeoglobus fulgidus.
Structure, 13, 2005
|
|
1E0K
 
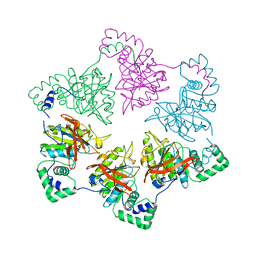 | | gp4d helicase from phage T7 | | Descriptor: | DNA HELICASE | | Authors: | Singleton, M.R, Sawaya, M.R, Ellenberger, T, Wigley, D.B. | | Deposit date: | 2000-03-30 | | Release date: | 2000-06-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal Structure of T7 Gene 4 Ring Helicase Indicates a Mechanism for Sequential Hydrolysis of Nucleotides
Cell(Cambridge,Mass.), 101, 2000
|
|
4ONK
 
 | | [Leu-5]-Enkephalin mutant - YVVFL | | Descriptor: | [Leu-5]-Enkephalin mutant - YVVFL | | Authors: | Sangwan, S, Eisenberg, D, Sawaya, M.R, Do, T.D, Bowers, M.T, Lapointe, N.E, Teplow, D.B, Feinstein, S.C. | | Deposit date: | 2014-01-28 | | Release date: | 2014-07-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Factors that drive Peptide assembly from native to amyloid structures: experimental and theoretical analysis of [leu-5]-enkephalin mutants.
J.Phys.Chem.B, 118, 2014
|
|
1EKF
 
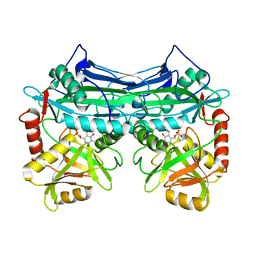 | | CRYSTALLOGRAPHIC STRUCTURE OF HUMAN BRANCHED CHAIN AMINO ACID AMINOTRANSFERASE (MITOCHONDRIAL) COMPLEXED WITH PYRIDOXAL-5'-PHOSPHATE AT 1.95 ANGSTROMS (ORTHORHOMBIC FORM) | | Descriptor: | BRANCHED CHAIN AMINO ACID AMINOTRANSFERASE (MITOCHONDRIAL), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Yennawar, N.H, Dunbar, J.H, Conway, M, Hutson, S.M, Farber, G.K. | | Deposit date: | 2000-03-08 | | Release date: | 2001-03-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structure of human mitochondrial branched-chain aminotransferase.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1EKP
 
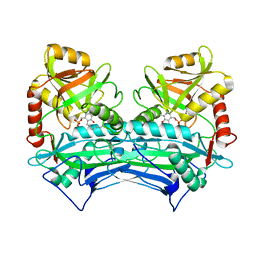 | | CRYSTAL STRUCTURE OF HUMAN BRANCHED CHAIN AMINO ACID AMINOTRANSFERASE (MITOCHONDRIAL) COMPLEXED WITH PYRIDOXAL-5'-PHOSPHATE AT 2.5 ANGSTROMS (MONOCLINIC FORM). | | Descriptor: | BRANCHED CHAIN AMINO ACID AMINOTRANSFERASE (MITOCHONDRIAL), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Yennawar, N.H, Dunbar, J.H, Conway, M, Hutson, S.M, Farber, G.K. | | Deposit date: | 2000-03-09 | | Release date: | 2001-03-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of human mitochondrial branched-chain aminotransferase.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1EKV
 
 | | HUMAN BRANCHED CHAIN AMINO ACID AMINOTRANSFERASE (MITOCHONDRIAL): THREE DIMENSIONAL STRUCTURE OF ENZYME INACTIVATED BY TRIS BOUND TO THE PYRIDOXAL-5'-PHOSPHATE ON ONE END AND ACTIVE SITE LYS202 NZ ON THE OTHER. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BRANCHED CHAIN AMINO ACID AMINOTRANSFERASE (MITOCHONDRIAL), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Yennawar, N.H, Dunbar, J.H, Conway, M, Hutson, S.M, Farber, G.K. | | Deposit date: | 2000-03-09 | | Release date: | 2001-03-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The structure of human mitochondrial branched-chain aminotransferase.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1F1F
 
 | | CRYSTAL STRUCTURE OF CYTOCHROME C6 FROM ARTHROSPIRA MAXIMA | | Descriptor: | CYTOCHROME C6, HEME C | | Authors: | Kerfeld, C.A, Serag, A.A, Sawaya, M.R, Krogmann, D.W, Yeates, T.O. | | Deposit date: | 2000-05-18 | | Release date: | 2001-08-08 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of cytochrome c-549 and cytochrome c6 from the cyanobacterium Arthrospira maxima.
Biochemistry, 40, 2001
|
|
1MY6
 
 | | The 1.6 A Structure of Fe-Superoxide Dismutase from the thermophilic cyanobacterium Thermosynechococcus elongatus : Correlation of EPR and Structural Characteristics | | Descriptor: | FE (III) ION, Iron (III) Superoxide Dismutase | | Authors: | Yoshida, S.M, Cascio, D, Sawaya, M.R, Yeates, T.O, Kerfeld, C.A. | | Deposit date: | 2002-10-03 | | Release date: | 2003-07-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The 1.6 A resolution structure of Fe-superoxide dismutase from the thermophilic cyanobacterium Thermosynechococcus elongatus.
J.BIOL.INORG.CHEM., 8, 2003
|
|
5VL4
 
 | | Accidental minimum contact crystal lattice formed by a redesigned protein oligomer | | Descriptor: | T33-53H-B | | Authors: | Cannon, K.A, Cascio, D, Sawaya, M.R, Park, R, Boyken, S, King, N, Yeates, T. | | Deposit date: | 2017-04-24 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Design and structure of two new protein cages illustrate successes and ongoing challenges in protein engineering.
Protein Sci., 29, 2020
|
|
