3T32
 
 | | Crystal structure of a putative C-S lyase from Bacillus anthracis | | Descriptor: | Aminotransferase, class I/II | | Authors: | Anderson, S.M, Wawrzak, Z, Gordon, E, Peterson, S.N, Porebski, P, Minor, W, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-07-24 | | Release date: | 2011-08-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a putative C-S lyase from Bacillus anthracis
TO BE PUBLISHED
|
|
4RPC
 
 | | Crystal structure of the putative alpha/beta hydrolase family protein from Desulfitobacterium hafniense | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, TETRAETHYLENE GLYCOL, putative alpha/beta hydrolase | | Authors: | Filippova, E.V, Wawrzak, Z, Minasov, G, Kiryukhina, O, Endres, M, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-10-30 | | Release date: | 2014-11-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the putative alpha/beta hydrolase family protein from Desulfitobacterium hafniense
To be Published
|
|
3Q7H
 
 | | Structure of the ClpP subunit of the ATP-dependent Clp Protease from Coxiella burnetii | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, CALCIUM ION, DI(HYDROXYETHYL)ETHER | | Authors: | Anderson, S.M, Wawrzak, Z, Gordon, E, Hasseman, J, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-01-04 | | Release date: | 2011-01-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the ClpP subunit of the ATP-dependent Clp Protease from Coxiella burnetii
To be Published
|
|
3QM3
 
 | | 1.85 Angstrom Resolution Crystal Structure of Fructose-bisphosphate Aldolase (Fba) from Campylobacter jejuni | | Descriptor: | CHLORIDE ION, FORMIC ACID, Fructose-bisphosphate aldolase, ... | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Onopriyenko, O, Papazisi, L, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-02-03 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1.85 Angstrom Resolution Crystal Structure of Fructose-bisphosphate Aldolase (Fba) from Campylobacter jejuni
TO BE PUBLISHED
|
|
4RSH
 
 | | Structure of a putative lipolytic protein of G-D-S-L family from Desulfitobacterium hafniense DCB-2 | | Descriptor: | CHLORIDE ION, Lipolytic protein G-D-S-L family | | Authors: | Filippova, E.V, Wawrzak, Z, Minasov, G, Kiryukhina, O, Endres, M, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-11-07 | | Release date: | 2014-11-19 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structure of a putative lipolytic protein of G-D-S-L family from Desulfitobacterium hafniense DCB-2
To be Published
|
|
4RS2
 
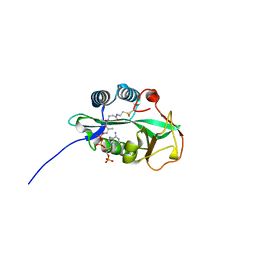 | | 1.55 Angstrom Crystal Structure of GNAT Family N-acetyltransferase (YhbS) from Escherichia coli in Complex with CoA | | Descriptor: | COENZYME A, Predicted acyltransferase with acyl-CoA N-acyltransferase domain | | Authors: | Minasov, G, Wawrzak, Z, Kuhn, M, Shuvalova, L, Dubrovska, I, Flores, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-11-06 | | Release date: | 2014-11-19 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | 1.55 Angstrom Crystal Structure of GNAT Family N-acetyltransferase (YhbS) from Escherichia coli in Complex with CoA.
TO BE PUBLISHED
|
|
3TYS
 
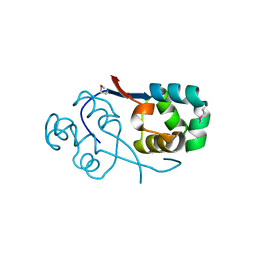 | | Crystal structure of transcriptional regulator VanUg, Form II | | Descriptor: | Predicted transcriptional regulator | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Depardieu, F, Courvalin, P, Shabalin, I, Chruszcz, M, Minor, W, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-26 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.121 Å) | | Cite: | Crystal structure of transcriptional regulator VanUg, Form II
TO BE PUBLISHED
|
|
3TYR
 
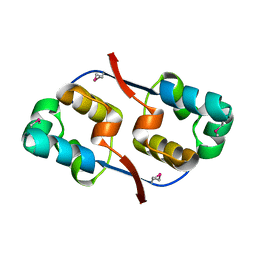 | | Crystal structure of transcriptional regulator VanUg, Form I | | Descriptor: | Transcriptional regulator | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Dong, A, Depardieu, F, Courvalin, P, Shabalin, I, Chruszcz, M, Minor, W, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-26 | | Release date: | 2011-10-12 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Crystal structure of transcriptional regulator VanUg, Form I
TO BE PUBLISHED
|
|
3R2U
 
 | | 2.1 Angstrom Resolution Crystal Structure of Metallo-beta-lactamase from Staphylococcus aureus subsp. aureus COL | | Descriptor: | CHLORIDE ION, FE (III) ION, MAGNESIUM ION, ... | | Authors: | Minasov, G, Wawrzak, Z, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Kiryukhina, O, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-03-14 | | Release date: | 2011-03-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 2.1 Angstrom Resolution Crystal Structure of Metallo-beta-lactamase Family Protein from Staphylococcus aureus subsp. aureus COL
TO BE PUBLISHED
|
|
3R8Y
 
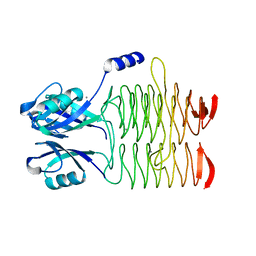 | | Structure of the Bacillus anthracis tetrahydropicolinate succinyltransferase | | Descriptor: | 2,3,4,5-tetrahydropyridine-2,6-dicarboxylate N-acetyltransferase, CALCIUM ION | | Authors: | Anderson, S.M, Wawrzak, Z, Onopriyenko, O, Peterson, S.N, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-03-24 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the Bacillus anthracis tetrahydropicolinate succinyltransferase
TO BE PUBLISHED
|
|
3UUW
 
 | | 1.63 Angstrom Resolution Crystal Structure of Dehydrogenase (MviM) from Clostridium difficile. | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Minasov, G, Wawrzak, Z, Kudritska, M, Grimshaw, S, Papazisi, L, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-11-28 | | Release date: | 2011-12-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | 1.63 Angstrom Resolution Crystal Structure of Dehydrogenase (MviM) from Clostridium difficile.
TO BE PUBLISHED
|
|
4W8K
 
 | | Crystal structure of a putative Cas1 enzyme from Vibrio phage ICP1 | | Descriptor: | Cas1 protein, POTASSIUM ION | | Authors: | Stogios, P.J, Wawrzak, Z, Onopriyeno, O, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-08-25 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | To be published
To Be Published
|
|
5KVK
 
 | | Crystal structure of the Competence-Damaged Protein (CinA) Superfamily Protein KP700603 from Klebsiella pneumoniae 700603 | | Descriptor: | Protein KP700603 | | Authors: | Stogios, P.J, Wawrzak, Z, Evdokimova, E, Di Leo, R, Grimshaw, S, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-14 | | Release date: | 2016-08-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | To be published
To Be Published
|
|
4CK8
 
 | | STEROL 14-ALPHA DEMETHYLASE (CYP51)FROM TRYPANOSOMA CRUZI IN COMPLEX WITH (R)-1-(2,4-dichlorophenyl)-2-(1H-imidazol-1-yl)ethyl 4-(4-(3,4- dichlorophenyl)piperazin-1-yl)phenylcarbamate (LFD) | | Descriptor: | (1R)-1-(2,4-dichlorophenyl)-2-(1H-imidazol-1-yl)ethyl {4-[4-(3,4-dichlorophenyl)piperazin-1-yl]phenyl}carbamate, PROTOPORPHYRIN IX CONTAINING FE, STEROL 14-ALPHA DEMETHYLASE | | Authors: | Friggeri, L, Wawrzak, Z, Tortorella, S, Lepesheva, G.I. | | Deposit date: | 2013-12-30 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural Basis for Rational Design of Inhibitors Targeting Trypanosoma Cruzi Sterol 14Alpha-Demethylase: Two Regions of the Enzyme Molecule Potentiate its Inhibition.
J.Med.Chem., 57, 2014
|
|
4CKA
 
 | | STEROL 14-ALPHA DEMETHYLASE (CYP51)FROM TRYPANOSOMA CRUZI IN COMPLEX WITH (S)-1-(4-fluorophenyl)-2-(1H-imidazol-1-yl)ethyl 4- isopropylphenylcarbamate (LFS) | | Descriptor: | (1S)-1-(4-fluorophenyl)-2-(1H-imidazol-1-yl)ethyl 4-isopropylphenylcarbamate, PROTOPORPHYRIN IX CONTAINING FE, STEROL 14-ALPHA DEMETHYLASE | | Authors: | Friggeri, L, Wawrzak, Z, Tortorella, S, Lepesheva, G.I. | | Deposit date: | 2013-12-31 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for Rational Design of Inhibitors Targeting Trypanosoma Cruzi Sterol 14Alpha-Demethylase: Two Regions of the Enzyme Molecule Potentiate its Inhibition.
J.Med.Chem., 57, 2014
|
|
4CK9
 
 | | STEROL 14-ALPHA DEMETHYLASE (CYP51)FROM TRYPANOSOMA CRUZI IN COMPLEX WITH (S)-1-(4-chlorophenyl)-2-(1H-imidazol-1-yl)ethyl 4- isopropylphenylcarbamate (LFT) | | Descriptor: | (1S)-1-(4-chlorophenyl)-2-(1H-imidazol-1-yl)ethyl [4-(propan-2-yl)phenyl]carbamate, PROTOPORPHYRIN IX CONTAINING FE, STEROL 14-ALPHA DEMETHYLASE | | Authors: | Friggeri, L, Wawrzak, Z, Tortorella, S, Lepesheva, G.I. | | Deposit date: | 2013-12-31 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structural Basis for Rational Design of Inhibitors Targeting Trypanosoma Cruzi Sterol 14Alpha-Demethylase: Two Regions of the Enzyme Molecule Potentiate its Inhibition.
J.Med.Chem., 57, 2014
|
|
4EG2
 
 | | 2.2 Angstrom Crystal Structure of Cytidine deaminase from Vibrio cholerae in Complex with Zinc and Uridine | | Descriptor: | ACETATE ION, Cytidine deaminase, MAGNESIUM ION, ... | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Wang, Y, Grimshaw, S, Papazisi, L, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-30 | | Release date: | 2012-05-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 2.2 Angstrom Crystal Structure of Cytidine deaminase from Vibrio cholerae in Complex with Zinc and Uridine.
TO BE PUBLISHED
|
|
4EHI
 
 | | An X-ray Crystal Structure of a putative Bifunctional Phosphoribosylaminoimidazolecarboxamide Formyltransferase/IMP Cyclohydrolase | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bifunctional purine biosynthesis protein PurH, SULFATE ION | | Authors: | Brunzelle, J.S, Wawrzak, Z, Onopriyenko, O, Kwok, J, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-02 | | Release date: | 2012-06-06 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | An X-ray Crystal Structure of a putative Bifunctional Phosphoribosylaminoimidazolecarboxamide Formyltransferase/IMP Cyclohydrolase
TO BE PUBLISHED
|
|
4EHJ
 
 | | An X-ray Structure of a Putative Phosphogylcerate Kinase from Francisella tularensis subsp. tularensis SCHU S4 | | Descriptor: | Phosphoglycerate kinase, SULFATE ION | | Authors: | Brunzelle, J.S, Wawrzak, Z, Skarina, T, Gordon, E, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-02 | | Release date: | 2012-05-09 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | An X-ray Structure of a Putative Phosphogylcerate Kinase from Francisella tularensis subsp. tularensis SCHU S4
To be Published
|
|
4EBJ
 
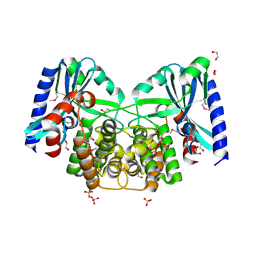 | | Crystal structure of aminoglycoside 4'-O-adenylyltransferase ANT(4')-IIb, apo | | Descriptor: | 1,2-ETHANEDIOL, Aminoglycoside nucleotidyltransferase, SULFATE ION | | Authors: | Stogios, P.J, Wawrzak, Z, Minasov, G, Evdokimova, E, Egorova, O, Yim, V, Kudritska, M, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-23 | | Release date: | 2012-04-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of aminoglycoside 4'-O-adenylyltransferase ANT(4')-IIb, apo
To be Published
|
|
4DD5
 
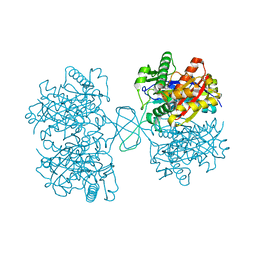 | | Biosynthetic Thiolase (ThlA1) from Clostridium difficile | | Descriptor: | Acetyl-CoA acetyltransferase | | Authors: | Filippova, E.V, Wawrzak, Z, Kudritska, M, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-01-18 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Biosynthetic Thiolase (ThlA1) from Clostridium difficile
To be Published
|
|
4EQB
 
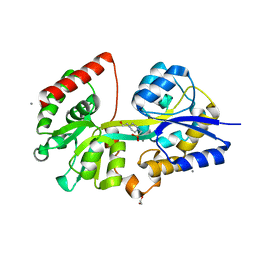 | | 1.5 Angstrom Crystal Structure of Spermidine/Putrescine ABC Transporter Substrate-Binding Protein PotD from Streptococcus pneumoniae strain Canada MDR_19A in Complex with Calcium and HEPES | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Minasov, G, Wawrzak, Z, Stogios, P.J, Kudritska, M, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-18 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.5 Angstrom Crystal Structure of Spermidine/Putrescine ABC Transporter Substrate-Binding Protein from Streptococcus pneumoniae strain Canada MDR_19A in Complex with Calcium and HEPES.
TO BE PUBLISHED
|
|
4EGU
 
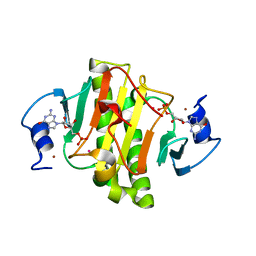 | | 0.95A Resolution Structure of a Histidine Triad Protein from Clostridium difficile | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, POTASSIUM ION, ZINC ION, ... | | Authors: | Anderson, S.M, Wawrzak, Z, Kudritska, M, Peterson, S.N, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-01 | | Release date: | 2012-04-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | 0.95A Resolution Structure of a Histidine Triad Protein from Clostridium difficile
To be Published
|
|
4E0B
 
 | | 2.17 Angstrom resolution crystal structure of malate dehydrogenase from Vibrio vulnificus CMCP6 | | Descriptor: | ACETATE ION, Malate dehydrogenase | | Authors: | Halavaty, A.S, Wawrzak, Z, Onopriyenko, O, Kwon, K, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-02 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | 2.17 Angstrom resolution crystal structure of malate dehydrogenase from Vibrio vulnificus CMCP6
To be Published
|
|
4EDP
 
 | | 1.85 Angstrom resolution crystal structure of an ABC transporter from Clostridium perfringens ATCC 13124 | | Descriptor: | ABC transporter, substrate-binding protein, ACETATE ION, ... | | Authors: | Halavaty, A.S, Wawrzak, Z, Onopriyenko, O, Peterson, S.N, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-27 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1.85 Angstrom resolution crystal structure of an ABC transporter from Clostridium perfringens ATCC 13124
To be Published
|
|
