2EKI
 
 | | Solution structures of the TGS domain of human developmentally-regulated GTP-binding protein 1 | | Descriptor: | Developmentally-regulated GTP-binding protein 1 | | Authors: | Sato, M, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-23 | | Release date: | 2007-09-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the TGS domain of human developmentally-regulated GTP-binding protein 1
To be Published
|
|
2ELI
 
 | | Solution structure of the second Phorbol esters/diacylglycerol binding domain of human Protein kinase C alpha type | | Descriptor: | Protein kinase C alpha type, ZINC ION | | Authors: | Tochio, N, Koshiba, S, Saito, K, Tomizawa, T, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-27 | | Release date: | 2008-04-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the second Phorbol esters/diacylglycerol binding domain of human Protein kinase C alpha type
To be Published
|
|
2DZL
 
 | | Solution Structure of the UBA domain in Human Protein FAM100B | | Descriptor: | Protein FAM100B | | Authors: | Zhao, C, Sato, M, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-09-29 | | Release date: | 2007-03-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the UBA domain in Human Protein FAM100B
To be Published
|
|
2EQJ
 
 | | Solution structure of the TUDOR domain of Metal-response element-binding transcription factor 2 | | Descriptor: | Metal-response element-binding transcription factor 2 | | Authors: | Dang, W, Muto, Y, Isono, K, Watanabe, S, Tarada, T, Kigawa, T, Koseki, H, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-30 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the TUDOR domain of Metal-response element-binding transcription factor 2
To be Published
|
|
7CGR
 
 | |
7CNQ
 
 | | Crystal structure of Agrobacterium tumefaciens aconitase X (holo-form) | | Descriptor: | (2~{S},3~{R})-3-oxidanylpyrrolidine-2-carboxylic acid, FE2/S2 (INORGANIC) CLUSTER, cis-3-hydroxy-L-proline dehydratase | | Authors: | Murase, Y, Watanabe, Y, Watanabe, S. | | Deposit date: | 2020-08-03 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of aconitase X enzymes from bacteria and archaea provide insights into the molecular evolution of the aconitase superfamily.
Commun Biol, 4, 2021
|
|
7CNP
 
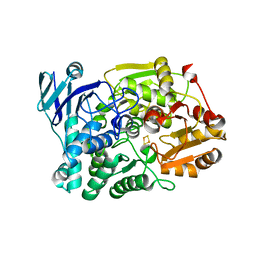 | |
7CNS
 
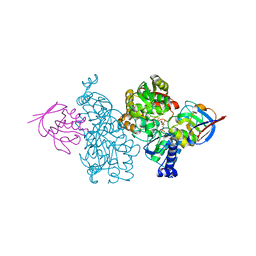 | | Crystal structure of Thermococcus kodakaraensis aconitase X (holo-form) | | Descriptor: | (3R)-3-HYDROXY-3-METHYL-5-(PHOSPHONOOXY)PENTANOIC ACID, DUF521 domain-containing protein, FE3-S4 CLUSTER, ... | | Authors: | Murase, Y, Watanabe, Y, Watanabe, S. | | Deposit date: | 2020-08-03 | | Release date: | 2021-06-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Crystal structures of aconitase X enzymes from bacteria and archaea provide insights into the molecular evolution of the aconitase superfamily.
Commun Biol, 4, 2021
|
|
7CNR
 
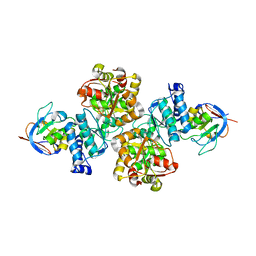 | |
7D2R
 
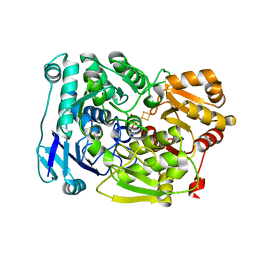 | | Crystal structure of Agrobacterium tumefaciens aconitase X mutant - S449C/C510V | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, SODIUM ION, ... | | Authors: | Murase, Y, Watanabe, Y, Watanabe, S. | | Deposit date: | 2020-09-17 | | Release date: | 2021-06-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Crystal structures of aconitase X enzymes from bacteria and archaea provide insights into the molecular evolution of the aconitase superfamily.
Commun Biol, 4, 2021
|
|
7DO6
 
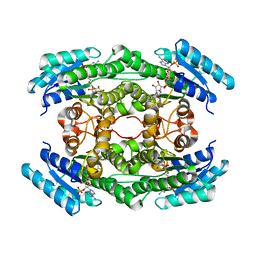 | |
7FGP
 
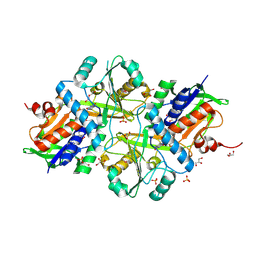 | |
7DO5
 
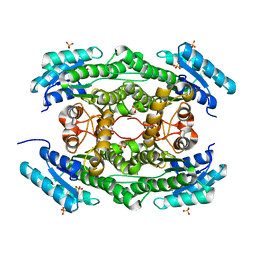 | |
7DO7
 
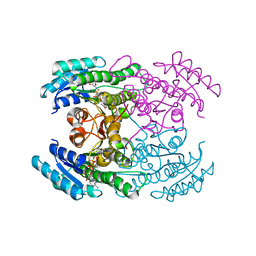 | |
7CGQ
 
 | |
2RR6
 
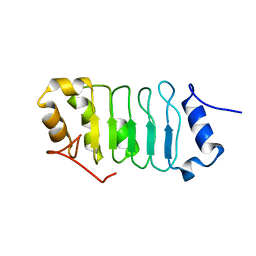 | | Solution structure of the leucine rich repeat of human acidic leucine-rich nuclear phosphoprotein 32 family member B | | Descriptor: | Acidic leucine-rich nuclear phosphoprotein 32 family member B | | Authors: | Tochio, N, Umehara, T, Tsuda, K, Koshiba, S, Harada, T, Watanabe, S, Tanaka, A, Kigawa, T, Yokoyama, S. | | Deposit date: | 2010-05-25 | | Release date: | 2010-06-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of histone chaperone ANP32B: interaction with core histones H3-H4 through its acidic concave domain.
J.Mol.Biol., 401, 2010
|
|
2RSX
 
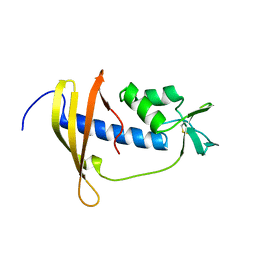 | | Solution structure of IseA, an inhibitor protein of DL-endopeptidases from Bacillus subtilis | | Descriptor: | Uncharacterized protein yoeB | | Authors: | Arai, R, Li, H, Tochio, N, Fukui, S, Kobayashi, N, Kitaura, C, Watanabe, S, Kigawa, T, Sekiguchi, J. | | Deposit date: | 2012-08-09 | | Release date: | 2012-10-31 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of IseA, an Inhibitor Protein of DL-Endopeptidases from Bacillus subtilis, Reveals a Novel Fold with a Characteristic Inhibitory Loop
J.Biol.Chem., 287, 2012
|
|
7DNN
 
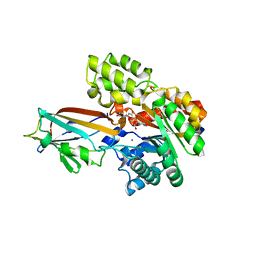 | | Crystal structure of the AgCarB2-C2 complex with homoorientin | | Descriptor: | 2-[3,4-bis(oxidanyl)phenyl]-6-[(2S,3R,4R,5S,6R)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]-5,7-bis(oxidanyl)chromen-4-one, AP_endonuc_2 domain-containing protein, AgCarC2, ... | | Authors: | Senda, M, Kumano, T, Watanabe, S, Kobayashi, M, Senda, T. | | Deposit date: | 2020-12-10 | | Release date: | 2021-10-20 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for the metabolism of xenobiotic C-glycosides by intestinal bacteria
Nat Commun, 2021
|
|
7DNM
 
 | | Crystal structure of the AgCarB2-C2 complex | | Descriptor: | AP_endonuc_2 domain-containing protein, AgCarC2, IODIDE ION, ... | | Authors: | Senda, M, Kumano, T, Watanabe, S, Kobayashi, M, Senda, T. | | Deposit date: | 2020-12-10 | | Release date: | 2021-10-20 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the metabolism of xenobiotic C-glycosides by intestinal bacteria
Nat Commun, 2021
|
|
7DVE
 
 | | Crystal structure of FAD-dependent C-glycoside oxidase | | Descriptor: | 6'''-hydroxyparomomycin C oxidase, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Senda, M, Watanabe, S, Kumano, T, Kobayashi, M, Senda, T. | | Deposit date: | 2021-01-13 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | FAD-dependent C -glycoside-metabolizing enzymes in microorganisms: Screening, characterization, and crystal structure analysis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CK5
 
 | | Solution structure of 28 amino acid polypeptide (354-381) in Plantago asiatica mosaic virus replicase bound to SDS micelle | | Descriptor: | PlAMV replicase peptide from RNA-dependent RNA polymerase | | Authors: | Komatsu, K, Sasaki, N, Yoshida, T, Suzuki, K, Masujima, Y, Hashimoto, M, Watanabe, S, Tochio, N, Kigawa, T, Yamaji, Y, Oshima, K, Namba, S, Nelson, R, Arie, T. | | Deposit date: | 2020-07-15 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Identification of a Proline-Kinked Amphipathic alpha-Helix Downstream from the Methyltransferase Domain of a Potexvirus Replicase and Its Role in Virus Replication and Perinuclear Complex Formation.
J.Virol., 95, 2021
|
|
7E7S
 
 | | WT transporter state1 | | Descriptor: | CALCIUM ION, Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 | | Authors: | Zhang, Y, Watanabe, S, Tsutsumi, A, Inaba, K. | | Deposit date: | 2021-02-27 | | Release date: | 2021-09-08 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM analysis provides new mechanistic insight into ATP binding to Ca 2+ -ATPase SERCA2b.
Embo J., 40, 2021
|
|
3VX3
 
 | | Crystal structure of [NiFe] hydrogenase maturation protein HypB from Thermococcus kodakarensis KOD1 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ATPase involved in chromosome partitioning, ... | | Authors: | Sasaki, D, Watanabe, S, Miki, K. | | Deposit date: | 2012-09-09 | | Release date: | 2013-02-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification and Structure of a Novel Archaeal HypB for [NiFe] Hydrogenase Maturation
J.Mol.Biol., 425, 2013
|
|
3WJQ
 
 | | Crystal structure of the HypE CN form | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, BENZAMIDINE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tominaga, T, Watanabe, S, Miki, K. | | Deposit date: | 2013-10-14 | | Release date: | 2013-12-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.645 Å) | | Cite: | Crystal structures of the carbamoylated and cyanated forms of HypE for [NiFe] hydrogenase maturation
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3WJP
 
 | | Crystal structure of the HypE CA form | | Descriptor: | BENZAMIDINE, GLYCEROL, Hydrogenase expression/formation protein HypE, ... | | Authors: | Tominaga, T, Watanabe, S, Miki, K. | | Deposit date: | 2013-10-14 | | Release date: | 2013-12-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.533 Å) | | Cite: | Crystal structures of the carbamoylated and cyanated forms of HypE for [NiFe] hydrogenase maturation
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
