1VB6
 
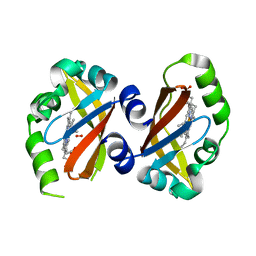 | | Crystal Structure of the heme PAS sensor domain of Ec DOS (oxygen-bound form) | | Descriptor: | Heme pas sensor protein, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kurokawa, H, Watanabe, M, Sagami, I, Mikami, B, Shimizu, T. | | Deposit date: | 2004-02-24 | | Release date: | 2005-04-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structure of oxygen-bound form of a Heme PAS domain of Ec DOS
To be Published
|
|
2CZW
 
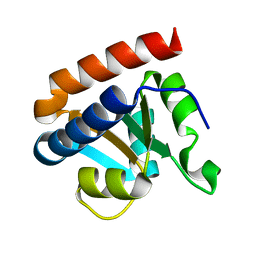 | | Crystal structure analysis of protein component Ph1496p of P.horikoshii ribonuclease P | | Descriptor: | 50S ribosomal protein L7Ae | | Authors: | Fukuhara, H, Kifusa, M, Watanabe, M, Terada, A, Honda, T, Numata, T, Kakuta, Y, Kimura, M. | | Deposit date: | 2005-07-19 | | Release date: | 2006-04-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A fifth protein subunit Ph1496p elevates the optimum temperature for the ribonuclease P activity from Pyrococcus horikoshii OT3
Biochem.Biophys.Res.Commun., 343, 2006
|
|
2DVY
 
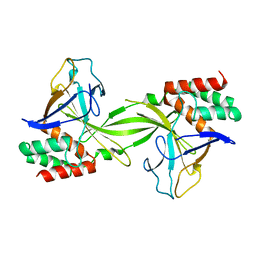 | | Crystal structure of restriction endonucleases PabI | | Descriptor: | Restriction endonuclease PabI | | Authors: | Miyazono, K, Watanabe, M, Kamo, M, Sawasaki, T, Nagata, K, Endo, Y, Tanokura, M, Kobayashi, I. | | Deposit date: | 2006-08-01 | | Release date: | 2007-05-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Novel protein fold discovered in the PabI family of restriction enzymes
Nucleic Acids Res., 35, 2007
|
|
5B5L
 
 | | Crystal structure of acetyl esterase mutant S10A with acetate ion | | Descriptor: | ACETATE ION, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Uechi, K, Kamachi, S, Akita, H, Mine, S, Watanabe, M. | | Deposit date: | 2016-05-12 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | crystal structure of acetyl esterase mutant S10A with acetate ion
To Be Published
|
|
7F8O
 
 | | Cryo-EM structure of the C-terminal deletion mutant of human PANX1 in a nanodisc | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Pannexin-1 | | Authors: | Kuzuya, M, Hirano, H, Hayashida, K, Watanabe, M, Kobayashi, K, Tani, K, Fujiyoshi, Y, Oshima, A. | | Deposit date: | 2021-07-02 | | Release date: | 2022-01-26 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of human pannexin-1 in nanodiscs reveal gating mediated by dynamic movement of the N terminus and phospholipids.
Sci.Signal., 15, 2022
|
|
7F8J
 
 | | Cryo-EM structure of human pannexin-1 in a nanodisc | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Pannexin-1 | | Authors: | Kuzuya, M, Hirano, H, Hayashida, K, Watanabe, M, Kobayashi, K, Tani, K, Fujiyoshi, Y, Oshima, A. | | Deposit date: | 2021-07-02 | | Release date: | 2022-01-26 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of human pannexin-1 in nanodiscs reveal gating mediated by dynamic movement of the N terminus and phospholipids.
Sci.Signal., 15, 2022
|
|
7F8N
 
 | | Human pannexin-1 showing a conformational change in the N-terminal domain and blocked pore | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Pannexin-1 | | Authors: | Kuzuya, M, Hirano, H, Hayashida, K, Watanabe, M, Kobayashi, K, Tani, K, Fujiyoshi, Y, Oshima, A. | | Deposit date: | 2021-07-02 | | Release date: | 2022-01-26 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of human pannexin-1 in nanodiscs reveal gating mediated by dynamic movement of the N terminus and phospholipids.
Sci.Signal., 15, 2022
|
|
7EAP
 
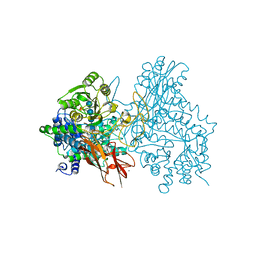 | | Crystal structure of IpeA-XXXG complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Matsuzawa, T, Watanabe, M, Nakamichi, Y, Akita, H, Yaoi, K. | | Deposit date: | 2021-03-08 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural basis for the catalytic mechanism of the glycoside hydrolase family 3 isoprimeverose-producing oligoxyloglucan hydrolase from Aspergillus oryzae.
Febs Lett., 596, 2022
|
|
3WQ8
 
 | | Monomer structure of hyperthermophilic beta-glucosidase mutant forming a dodecameric structure in the crystal form | | Descriptor: | Beta-glucosidase | | Authors: | Nakabayashi, M, Kataoka, M, Watanabe, M, Ishikawa, K. | | Deposit date: | 2014-01-23 | | Release date: | 2014-07-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Monomer structure of a hyperthermophilic beta-glucosidase mutant forming a dodecameric structure in the crystal form.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
3WO8
 
 | | Crystal structure of the beta-N-acetylglucosaminidase from Thermotoga maritima | | Descriptor: | Beta-N-acetylglucosaminidase | | Authors: | Mine, S, Kado, Y, Watanabe, M, Inoue, T, Ishikawa, K. | | Deposit date: | 2013-12-20 | | Release date: | 2014-12-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | The structure of hyperthermophilic beta-N-acetylglucosaminidase reveals a novel dimer architecture associated with the active site.
Febs J., 281, 2014
|
|
2YU0
 
 | | Solution structures of the PAAD_DAPIN domain of mus musculus interferon-activatable protein 205 | | Descriptor: | Interferon-activable protein 205 | | Authors: | Sato, M, Tochio, N, Koshiba, S, Watanabe, M, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the PAAD_DAPIN domain of mus musculus interferon-activatable protein 205
To be Published
|
|
3AQD
 
 | |
3AK0
 
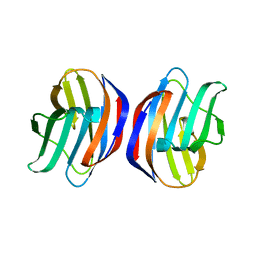 | | Crystal Structure of Ancestral Congerin Con-anc'-N28K | | Descriptor: | Ancestral congerin Con-anc, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Konno, A, Kitagawa, A, Watanabe, M, Ogawa, T, Shirai, T. | | Deposit date: | 2010-06-29 | | Release date: | 2011-05-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Tracing protein evolution through ancestral structures of fish galectin
Structure, 19, 2011
|
|
3AJY
 
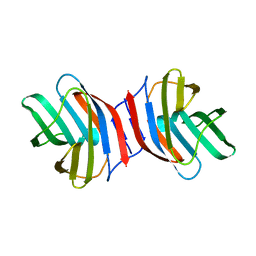 | | Crystal Structure of Ancestral Congerin Con-anc | | Descriptor: | Ancestral congerin Con-anc, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Konno, A, Kitagawa, A, Watanabe, M, Ogawa, T, Shirai, T. | | Deposit date: | 2010-06-29 | | Release date: | 2011-05-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Tracing protein evolution through ancestral structures of fish galectin
Structure, 19, 2011
|
|
3AJZ
 
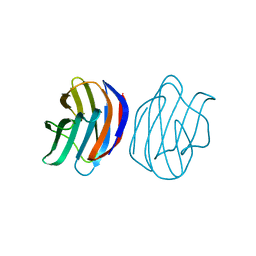 | | Crystal Structure of Ancestral Congerin Con-anc | | Descriptor: | Ancestral congerin Con-anc, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Konno, A, Kitagawa, A, Watanabe, M, Ogawa, T, Shirai, T. | | Deposit date: | 2010-06-29 | | Release date: | 2011-05-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Tracing protein evolution through ancestral structures of fish galectin
Structure, 19, 2011
|
|
3ST7
 
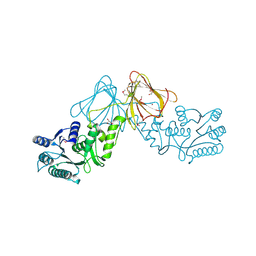 | | Crystal Structure of capsular polysaccharide assembling protein CapF from staphylococcus aureus | | Descriptor: | Capsular polysaccharide synthesis enzyme Cap5F, GLYCEROL, ZINC ION | | Authors: | Miyafusa, T, Tanaka, Y, Kuroda, M, Yao, M, Watanabe, M, Ohta, T, Tanaka, I, Caaveiro, J.M.M, Tsumoto, K. | | Deposit date: | 2011-07-09 | | Release date: | 2012-02-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of the enzyme CapF of Staphylococcus aureus reveals a unique architecture composed of two functional domains.
Biochem.J., 443, 2012
|
|
