3CU5
 
 | | Crystal structure of a two component transcriptional regulator AraC from Clostridium phytofermentans ISDg | | Descriptor: | Two component transcriptional regulator, AraC family | | Authors: | Malashkevich, V.N, Toro, R, Wasserman, S.R, Meyer, A, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-15 | | Release date: | 2008-05-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a two component transcriptional regulator AraC from Clostridium phytofermentans ISDg.
To be Published
|
|
4RXT
 
 | | Crystal structure of carbohydrate transporter solute binding protein Arad_9553 from Agrobacterium Radiobacter, Target EFI-511541, in complex with D-arabinose | | Descriptor: | Sugar ABC transporter, alpha-L-arabinopyranose | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Hillerich, B, Siedel, R.D, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-12-11 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal Structure of Sugar Binding Transporter Arad_9553 from Agrobacterium Radiobacter, Target Efi-511541
To be Published
|
|
3CFY
 
 | | Crystal structure of signal receiver domain of putative Luxo repressor protein from Vibrio parahaemolyticus | | Descriptor: | Putative LuxO repressor protein | | Authors: | Patskovsky, Y, Ramagopal, U.A, Fong, R, Freeman, J, Iizuka, M, Groshong, C, Smith, D, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-03-04 | | Release date: | 2008-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Signal Receiver Domain of Putative Luxo Repressor Protein from Vibrio Parahaemolyticus.
To be Published
|
|
3CYJ
 
 | | Crystal structure of a mandelate racemase/muconate lactonizing enzyme-like protein from Rubrobacter xylanophilus | | Descriptor: | GLYCEROL, Mandelate racemase/muconate lactonizing enzyme-like protein, SODIUM ION | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Zhang, F, Bravo, J, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-25 | | Release date: | 2008-05-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a mandelate racemase/muconate lactonizing enzyme-like protein from Rubrobacter xylanophilus.
To be Published
|
|
3CTA
 
 | | Crystal structure of riboflavin kinase from Thermoplasma acidophilum | | Descriptor: | Riboflavin kinase | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Mendoza, M, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-11 | | Release date: | 2008-04-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of riboflavin kinase from Thermoplasma acidophilum.
To be Published
|
|
3CTP
 
 | | Crystal structure of periplasmic binding protein/LacI transcriptional regulator from Alkaliphilus metalliredigens QYMF complexed with D-xylulofuranose | | Descriptor: | Periplasmic binding protein/LacI transcriptional regulator, SODIUM ION, beta-D-xylulofuranose | | Authors: | Malashkevich, V.N, Toro, R, Wasserman, S.R, Meyer, A, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-14 | | Release date: | 2008-05-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Crystal structure of periplasmic binding protein/LacI transcriptional regulator from Alkaliphilus metalliredigens QYMF complexed with L-xylulose.
To be Published
|
|
3EYE
 
 | | Crystal structure of PTS system N-acetylgalactosamine-specific IIB component 1 from Escherichia coli | | Descriptor: | PTS system N-acetylgalactosamine-specific IIB component 1 | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Do, J, Sampathkumar, P, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-10-20 | | Release date: | 2008-11-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of PTS system N-acetylgalactosamine-specific IIB component 1 from Escherichia coli
To be Published
|
|
3F13
 
 | | Crystal structure of putative nudix hydrolase family member from Chromobacterium violaceum | | Descriptor: | putative nudix hydrolase family member | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Do, J, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-10-27 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of putative nudix hydrolase family member from Chromobacterium violaceum
To be Published
|
|
3F6C
 
 | | CRYSTAL STRUCTURE OF N-TERMINAL DOMAIN OF POSITIVE TRANSCRIPTION REGULATOR evgA FROM ESCHERICHIA COLI | | Descriptor: | GLYCEROL, Positive transcription regulator evgA | | Authors: | Patskovsky, Y, Romero, R, Freeman, J, Wu, B, Bain, K, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-11-05 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | CRYSTAL STRUCTURE OF N-TERMINAL DOMAIN OF POSITIVE TRANSCRIPTION REGULATOR evgA FROM ESCHERICHIA COLI
To be Published
|
|
3EQZ
 
 | | Crystal structure of a response regulator from Colwellia psychrerythraea | | Descriptor: | Response regulator | | Authors: | Bonanno, J.B, Gilmore, M, Bain, K.T, Iizuka, M, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-10-01 | | Release date: | 2008-10-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of a response regulator from Colwellia psychrerythraea
To be Published
|
|
3EVT
 
 | | Crystal structure of phosphoglycerate dehydrogenase from Lactobacillus plantarum | | Descriptor: | Phosphoglycerate dehydrogenase | | Authors: | Bonanno, J.B, Gilmore, M, Bain, K.T, Do, J, Sampathkumar, P, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-10-13 | | Release date: | 2008-10-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of phosphoglycerate dehydrogenase from Lactobacillus plantarum
To be Published
|
|
3DFH
 
 | | crystal structure of putative mandelate racemase / muconate lactonizing enzyme from Vibrionales bacterium SWAT-3 | | Descriptor: | SODIUM ION, mandelate racemase | | Authors: | Malashkevich, V.N, Toro, R, Wasserman, S.R, Meyer, A.J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-06-12 | | Release date: | 2008-07-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | crystal structure of putative mandelate racemase / muconate lactonizing enzyme from Vibrionales bacterium SWAT-3
To be Published
|
|
3FBG
 
 | | Crystal structure of a putative arginate lyase from Staphylococcus haemolyticus | | Descriptor: | MAGNESIUM ION, putative arginate lyase | | Authors: | Bonanno, J.B, Gilmore, M, Bain, K.T, Do, J, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-11-19 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a putative arginate lyase from Staphylococcus haemolyticus
To be Published
|
|
2PMQ
 
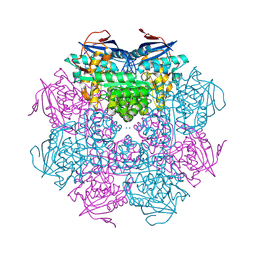 | | Crystal structure of a mandelate racemase/muconate lactonizing enzyme from Roseovarius sp. HTCC2601 | | Descriptor: | MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Lau, C, Sridhar, V, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-04-23 | | Release date: | 2007-05-08 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery of new enzymes and metabolic pathways by using structure and genome context.
Nature, 502, 2013
|
|
3C19
 
 | | Crystal structure of protein MK0293 from Methanopyrus kandleri AV19 | | Descriptor: | GLYCEROL, PHOSPHATE ION, Uncharacterized protein MK0293 | | Authors: | Patskovsky, Y, Romero, R, Bonanno, J.B, Malashkevich, V, Dickey, M, Chang, S, Koss, J, Bain, K, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-01-22 | | Release date: | 2008-02-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of protein MK0293 from Methanopyrus kandleri AV19.
To be Published
|
|
3C97
 
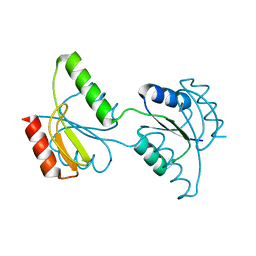 | | Crystal structure of the response regulator receiver domain of a signal transduction histidine kinase from Aspergillus oryzae | | Descriptor: | Signal transduction histidine kinase | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Chang, S, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-02-15 | | Release date: | 2008-03-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the response regulator receiver domain of a signal transduction histidine kinase from Aspergillus oryzae.
To be Published
|
|
3BY5
 
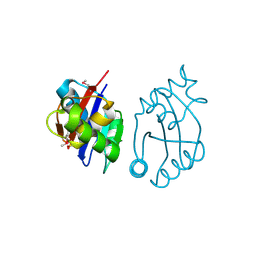 | | Crystal structure of cobalamin biosynthesis protein chiG from Agrobacterium tumefaciens str. C58 | | Descriptor: | Cobalamin biosynthesis protein, SULFATE ION | | Authors: | Patskovsky, Y, Bonanno, J.B, Sojitra, S, Rutter, M, Iizuka, M, Maletic, M, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-01-15 | | Release date: | 2008-01-22 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Crystal structure of cobalamin biosynthesis protein from Agrobacterium tumefaciens str. C58.
To be Published
|
|
3BMA
 
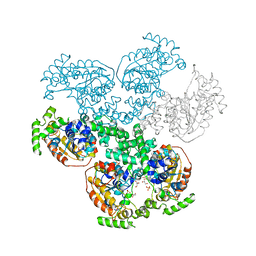 | | Crystal structure of D-alanyl-lipoteichoic acid synthetase from Streptococcus pneumoniae R6 | | Descriptor: | D-alanyl-lipoteichoic acid synthetase, GLYCEROL, SULFATE ION | | Authors: | Patskovsky, Y, Sridhar, V, Bonanno, J.B, Smith, D, Rutter, M, Iizuka, M, Koss, J, Bain, K, Gheyi, T, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-12-12 | | Release date: | 2007-12-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal Structure of probable D-Alanyl-Lipoteichoic Acid Synthetase from Streptococcus pneumoniae.
To be Published
|
|
3BQ9
 
 | | Crystal structure of predicted nucleotide-binding protein from Idiomarina baltica OS145 | | Descriptor: | GLYCEROL, Predicted Rossmann fold nucleotide-binding domain-containing protein, SULFATE ION | | Authors: | Patskovsky, Y, Toro, R, Meyer, A.J, Dickey, M, Eberle, M, Koss, J, Groshong, C, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-12-19 | | Release date: | 2008-01-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Predicted Nucleotide-Binding Protein from Idiomarina baltica.
To be Published
|
|
3E61
 
 | | Crystal structure of a putative transcriptional repressor of ribose operon from Staphylococcus saprophyticus subsp. saprophyticus | | Descriptor: | GLYCEROL, Putative transcriptional repressor of ribose operon | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Chang, S, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-08-14 | | Release date: | 2008-08-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a putative transcriptional repressor of ribose operon from Staphylococcus saprophyticus subsp. saprophyticus
To be Published
|
|
3E9N
 
 | | Crystal structure of a putative short-chain dehydrogenase/reductase from Corynebacterium glutamicum | | Descriptor: | PUTATIVE SHORT-CHAIN DEHYDROGENASE/REDUCTASE | | Authors: | Bonanno, J.B, Gilmore, M, Bain, K.T, Hu, S, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-08-22 | | Release date: | 2008-09-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a putative short-chain dehydrogenase/reductase from Corynebacterium glutamicum
To be Published
|
|
4RU1
 
 | | Crystal structure of carbohydrate transporter ACEI_1806 from Acidothermus cellulolyticus 11B, TARGET EFI-510965, in complex with myo-inositol | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, CITRIC ACID, Monosaccharide ABC transporter substrate-binding protein, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Hillerich, B, Siede, R.D, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-11-17 | | Release date: | 2014-12-17 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of carbohydrate transporter ACEI_1806 from Acidothermus cellulolyticus, TARGET EFI-510965.
To be Published
|
|
3E3M
 
 | | Crystal structure of a LacI family transcriptional regulator from Silicibacter pomeroyi | | Descriptor: | Transcriptional regulator, LacI family | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Iizuka, M, Ozyurt, S, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-08-07 | | Release date: | 2008-08-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a LacI family transcriptional regulator from Silicibacter pomeroyi
To be Published
|
|
4WUT
 
 | | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM AGROBACTERIUM VITIS (Avi_5133, TARGET EFI-511220) WITH BOUND D-FUCOSE | | Descriptor: | ABC transporter substrate binding protein (Ribose), CALCIUM ION, CHLORIDE ION, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-11-03 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM AGROBACTERIUM VITIS (Avi_5133, TARGET EFI-511220) WITH BOUND D-FUCOSE
To be published
|
|
3EDM
 
 | | Crystal structure of a short chain dehydrogenase from Agrobacterium tumefaciens | | Descriptor: | Short chain dehydrogenase | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Chang, S, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-09-03 | | Release date: | 2008-09-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a short chain dehydrogenase from Agrobacterium tumefaciens
To be Published
|
|
