3V3W
 
 | | Crystal structure of an enolase from the soil bacterium Cellvibrio japonicus (TARGET EFI-502161) with bound MG and glycerol | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Sojitra, S, Seidel, R, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-12-14 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of an enolase from the soil bacterium Cellvibrio japonicus (TARGET EFI-502161) with bound MG and glycerol
to be published
|
|
3V4B
 
 | | Crystal structure of an enolase from the soil bacterium Cellvibrio japonicus (TARGET EFI-502161) with bound MG and L-tartrate | | Descriptor: | CHLORIDE ION, L(+)-TARTARIC ACID, MAGNESIUM ION, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Sojitra, S, Seidel, R.D, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Al Obaidi, N, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-12-14 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of an enolase from the soil bacterium Cellvibrio japonicus (TARGET EFI-502161) with bound MG and l-tartrate
to be published
|
|
3T6C
 
 | | Crystal structure of an enolase from pantoea ananatis (efi target efi-501676) with bound d-gluconate and mg | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, D-gluconic acid, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-07-28 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Crystal structure of an enolase from pantoea ananatis (efi target efi-501676) with bound d-gluconate and mg
to be published
|
|
5HFK
 
 | | CRYSTAL STRUCTURE OF A GLUTATHIONE S-TRANSFERASE PROTEIN FROM ESCHERICHIA COLI OCh 157:H7 STR. SAKAI (ECs3186, TARGET EFI-507414) WITH BOUND GLUTATHIONE | | Descriptor: | Disulfide-bond oxidoreductase YfcG, GLUTATHIONE | | Authors: | Himmel, D.M, Toro, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI), New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2016-01-07 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.551 Å) | | Cite: | CRYSTAL STRUCTURE OF A GLUTATHIONE S-TRANSFERASE PROTEIN FROM ESCHERICHIA COLI OCh 157:H7 STR. SAKAI (ECs3186, TARGET EFI-507414) WITH BOUND GLUTATHIONE
TO BE PUBLISHED
|
|
6CQA
 
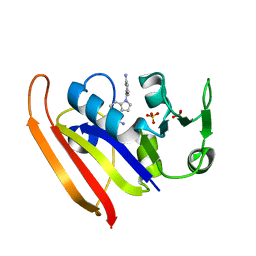 | | E. coli DHFR complex with inhibitor AMPQD | | Descriptor: | 7-[(3-aminophenyl)methyl]-7H-pyrrolo[3,2-f]quinazoline-1,3-diamine, Dihydrofolate reductase, SULFATE ION | | Authors: | Cao, H, Rodrigues, J, Benach, J, Wasserman, S, Morisco, L, Koss, J, Shakhnovich, E, Skolnick, J. | | Deposit date: | 2018-03-14 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of a tetrahydrofolate-bound dihydrofolate reductase reveals the origin of slow product release.
Commun Biol, 1, 2018
|
|
3THU
 
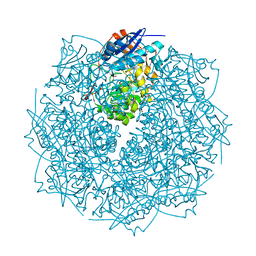 | | Crystal structure of an enolase from sphingomonas sp. ska58 (efi target efi-501683) with bound mg | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-08-19 | | Release date: | 2011-09-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of an enolase from sphingomonas sp. ska58 (efi target efi-501683) with bound mg
to be published
|
|
3UGV
 
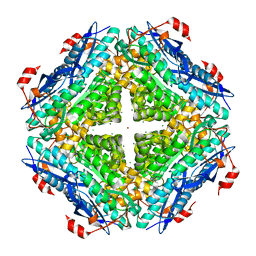 | | Crystal structure of an enolase from alpha pretobacterium bal199 (EFI TARGET EFI-501650) with bound MG | | Descriptor: | CHLORIDE ION, Enolase, MAGNESIUM ION, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-11-03 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of an enolase from alpha pretobacterium bal199 (EFI TARGET EFI-501650) with bound MG
to be published
|
|
3TJI
 
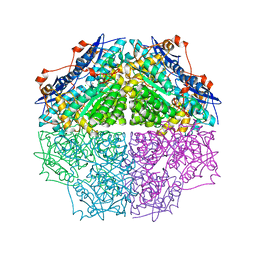 | | CRYSTAL STRUCTURE OF AN ENOLASE FROM ENTEROBACTER sp. 638 (EFI TARGET EFI-501662) with BOUND MG | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-08-24 | | Release date: | 2011-09-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of an enolase from enterobacter sp. 638 (efi target efi-501662) with boung mg
to be published
|
|
3C6F
 
 | | Crystal structure of protein Bsu07140 from Bacillus subtilis | | Descriptor: | GLYCEROL, YetF protein | | Authors: | Patskovsky, Y, Min, T, Zhang, A, Adams, J, Groshong, C, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-02-04 | | Release date: | 2008-02-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of protein Bsu07140 from Bacillus subtilis.
To be Published
|
|
3BT5
 
 | | Crystal structure of DUF305 fragment from Deinococcus radiodurans | | Descriptor: | CHLORIDE ION, Uncharacterized protein DUF305 | | Authors: | Ramagopal, U.A, Patskovsky, Y, Rutter, M, Toro, R, Bain, K, Meyer, A.J, Powell, A, Gheyi, T, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-12-27 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of DUF305 fragment from Deinococcus radiodurans.
To be Published
|
|
3BVC
 
 | | Crystal structure of uncharacterized protein Ism_01780 from Roseovarius nubinhibens ISM | | Descriptor: | CALCIUM ION, NICKEL (II) ION, Uncharacterized protein Ism_01780 | | Authors: | Patskovsky, Y, Toro, R, Meyer, A.J, Rutter, M, Iizuka, M, Maletic, M, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-01-06 | | Release date: | 2008-02-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of an uncharacterized protein Ism_01780 from Roseovarius nubinhibens.
To be Published
|
|
3L49
 
 | | CRYSTAL STRUCTURE OF ABC SUGAR TRANSPORTER SUBUNIT FROM Rhodobacter sphaeroides 2.4.1 | | Descriptor: | ABC sugar (Ribose) transporter, periplasmic substrate-binding subunit, UNKNOWN LIGAND | | Authors: | Patskovsky, Y, Ozyurt, S, Dickey, M, Do, J, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York Structural GenomiX Research Consortium (NYSGXRC), New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-12-18 | | Release date: | 2010-01-05 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | CRYSTAL STRUCTURE OF ABC SUGAR TRANSPORTER FROM Rhodobacter sphaeroides
To be Published
|
|
3EOI
 
 | | CRYSTAL STRUCTURE OF putative PROTEIN PilM from Escherichia coli B7A | | Descriptor: | PilM | | Authors: | Malashkevich, V.N, Toro, R, Bonanno, J.B, Sauder, J.M, Wasserman, S, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-09-26 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal structure of an uncharacterized protein
to be published
|
|
4RXU
 
 | | Crystal structure of carbohydrate transporter solute binding protein CAUR_1924 from Chloroflexus aurantiacus, Target EFI-511158, in complex with D-glucose | | Descriptor: | CITRIC ACID, Periplasmic sugar-binding protein, beta-D-glucopyranose | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Hillerich, B, Siedel, R.D, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-12-11 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of sugar transporter CAUR_1924 from Chloroflexus aurantiacus, Target EFI-511158
To be Published
|
|
4RY1
 
 | | Crystal structure of periplasmic solute binding protein ECA2210 from Pectobacterium atrosepticum SCRI1043, Target EFI-510858 | | Descriptor: | ACETATE ION, GLYCEROL, Periplasmic solute binding protein | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Hillerich, B, Siedel, R.D, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-12-12 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of periplasmic solute binding protein ECA2210 from Pectobacterium atrosepticum, Target EFI-510858
To be Published
|
|
4RXL
 
 | | Crystal structure of Molybdenum ABC transporter solute binding protein Vc_A0726 from Vibrio Cholerae, Target EFI-510913, in complex with tungstate | | Descriptor: | Molybdenum ABC transporter, periplasmic molybdenum-binding protein, TUNGSTATE(VI)ION | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Hillerich, B, Siedel, R.D, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-12-11 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal Structure of Molybdate Transporter Vc_A0726 from Vibrio Cholerae, Target EFI-510913
To be Published
|
|
4RY9
 
 | | Crystal structure of carbohydrate transporter solute binding protein VEIS_2079 from Verminephrobacter eiseniae EF01-2, TARGET EFI-511009, a complex with D-TALITOL | | Descriptor: | BICARBONATE ION, D-altritol, GLYCEROL, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Hillerich, B, Siedel, R.D, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-12-13 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of carbohydrate transporter solute binding protein VEIS_2079 from Verminephrobacter eiseniae
To be Published
|
|
4RYA
 
 | | Crystal structure of abc transporter solute binding protein AVI_3567 from AGROBACTERIUM VITIS S4, TARGET EFI-510645, with bound D-mannitol | | Descriptor: | ABC transporter substrate binding protein (Sorbitol), ACETATE ION, D-MANNITOL | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Hillerich, B, Siedel, R.D, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-12-13 | | Release date: | 2014-12-24 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Periplasmic Solute-Binding Protein Avi_3567 from Agrobacterium Vitis, Target Efi-510645
To be Published
|
|
3CBW
 
 | | Crystal structure of the YdhT protein from Bacillus subtilis | | Descriptor: | CITRIC ACID, YdhT protein | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Iizuka, M, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-02-23 | | Release date: | 2008-03-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.269 Å) | | Cite: | Crystal structure of the YdhT protein from Bacillus subtilis.
To be Published
|
|
3CDX
 
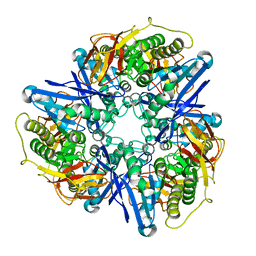 | | Crystal structure of succinylglutamatedesuccinylase/aspartoacylase from Rhodobacter sphaeroides | | Descriptor: | CALCIUM ION, Succinylglutamatedesuccinylase/aspartoacylase | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Iizuka, M, Patterson, K, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-02-27 | | Release date: | 2008-03-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of succinylglutamatedesuccinylase/aspartoacylase from Rhodobacter sphaeroides.
To be Published
|
|
4RXM
 
 | | Crystal structure of periplasmic ABC transporter solute binding protein A7JW62 from Mannheimia haemolytica PHL213, Target EFI-511105, in complex with Myo-inositol | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, ASPARTIC ACID, CHLORIDE ION, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Hillerich, B, Siedel, R.D, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-12-11 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of solute binding sugar transporter A7JW62 from Mannheimia haemolytica, Target EFI-511105.
To be Published
|
|
3C9F
 
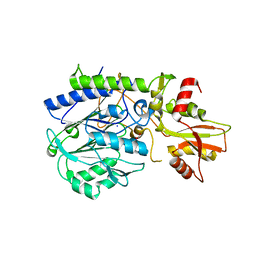 | | Crystal structure of 5'-nucleotidase from Candida albicans SC5314 | | Descriptor: | 5'-nucleotidase, FORMIC ACID, SODIUM ION, ... | | Authors: | Patskovsky, Y, Romero, R, Gilmore, M, Eberle, M, Bain, K, Smith, D, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-02-15 | | Release date: | 2008-02-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of 5'-nucleotidase from Candida albicans.
To be Published
|
|
4RY0
 
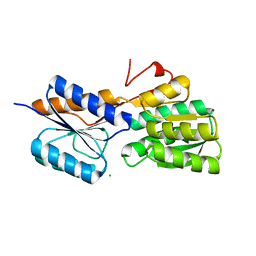 | | Crystal structure of ribose transporter solute binding protein RHE_PF00037 from Rhizobium etli CFN 42, TARGET EFI-511357, in complex with D-ribose | | Descriptor: | CALCIUM ION, CHLORIDE ION, Probable ribose ABC transporter, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Hillerich, B, Siedel, R.D, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-12-12 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of Ribose Transporter Solute Binding Protein from Rhizobium Etly, Target Efi-511357
To be Published
|
|
3L8K
 
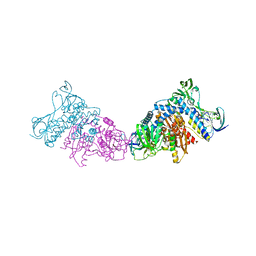 | | Crystal structure of a dihydrolipoyl dehydrogenase from Sulfolobus solfataricus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Dihydrolipoyl dehydrogenase, PHOSPHATE ION | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Miller, S, Sampathkumar, P, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-12-31 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a dihydrolipoyl dehydrogenase from Sulfolobus solfataricus
To be Published
|
|
3CZ5
 
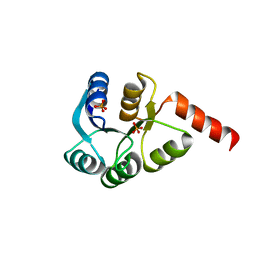 | | Crystal structure of two-component response regulator, LuxR family, from Aurantimonas sp. SI85-9A1 | | Descriptor: | PHOSPHATE ION, Two-component response regulator, LuxR family | | Authors: | Malashkevich, V.N, Toro, R, Wasserman, S.R, Meyer, A, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-28 | | Release date: | 2008-05-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of two-component response regulator, LuxR family, from Aurantimonas sp. SI85-9A1.
To be Published
|
|
