1EP0
 
 | | HIGH RESOLUTION CRYSTAL STRUCTURE OF DTDP-6-DEOXY-D-XYLO-4-HEXULOSE 3,5-EPIMERASE FROM METHANOBACTERIUM THERMOAUTOTROPHICUM | | Descriptor: | DTDP-6-DEOXY-D-XYLO-4-HEXULOSE 3,5-EPIMERASE | | Authors: | Christendat, D, Saridakis, V, Bochkarev, A, Pai, E.F, Arrowsmith, C.H, Edwards, A.M, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2000-03-24 | | Release date: | 2000-12-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of dTDP-4-keto-6-deoxy-D-hexulose 3,5-epimerase from Methanobacterium thermoautotrophicum complexed with dTDP.
J.Biol.Chem., 275, 2000
|
|
1EPZ
 
 | | CRYSTAL STRUCTURE OF DTDP-6-DEOXY-D-XYLO-4-HEXULOASE 3,5-EPIMERASE FROM METHANOBACTERIUM THERMOAUTOTROPHICUM WITH BOUND LIGAND. | | Descriptor: | DTDP-6-DEOXY-D-XYLO-4-HEXULOSE 3,5-EPIMERASE, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Christendat, D, Saridakis, V, Bochkarev, A, Pai, E.F, Arrowsmith, C, Edwards, A.M. | | Deposit date: | 2000-03-30 | | Release date: | 2000-12-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of dTDP-4-keto-6-deoxy-D-hexulose 3,5-epimerase from Methanobacterium thermoautotrophicum complexed with dTDP.
J.Biol.Chem., 275, 2000
|
|
1EJ2
 
 | | Crystal structure of methanobacterium thermoautotrophicum nicotinamide mononucleotide adenylyltransferase with bound NAD+ | | Descriptor: | NICOTINAMIDE MONONUCLEOTIDE ADENYLYLTRANSFERASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Saridakis, V, Christendat, D, Kimber, M.S, Edwards, A.M, Pai, E.F, Midwest Center for Structural Genomics (MCSG), Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2000-02-29 | | Release date: | 2001-03-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into ligand binding and catalysis of a central step in NAD+ synthesis: structures of Methanobacterium thermoautotrophicum NMN adenylyltransferase complexes.
J.Biol.Chem., 276, 2001
|
|
7N27
 
 | | Crystal Structure of chromodomain of CDYL in complex with inhibitor UNC6261 | | Descriptor: | Isoform 2 of Chromodomain Y-like protein, SODIUM ION, UNKNOWN ATOM OR ION, ... | | Authors: | Beldar, S, Dong, A, Loppnau, P, Min, J, Arrowsmith, C.H, Edwards, A.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-05-28 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of chromodomain of CDYL in complex with inhibitor UNC6261
To Be Published
|
|
6XKB
 
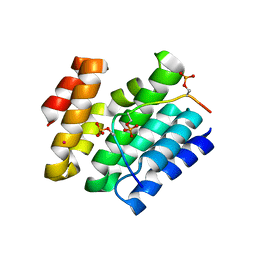 | | Crystal structure of SR-related and CTD-associated factor 4(SCAF4-CID)with peptide S2,S5p-CTD | | Descriptor: | S2,S5p-CTD peptide, SR-related and CTD-associated factor 4, UNKNOWN ATOM OR ION | | Authors: | Zhou, M.Q, Dong, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-26 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the recognition of the S2, S5-phosphorylated RNA polymerase II CTD by the mRNA anti-terminator protein hSCAF4.
Febs Lett., 596, 2022
|
|
1MXI
 
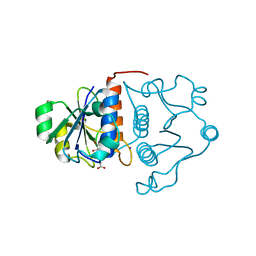 | | Structure of YibK from Haemophilus influenzae (HI0766): a Methyltransferase with a Cofactor Bound at a Site Formed by a Knot | | Descriptor: | Hypothetical tRNA/rRNA methyltransferase HI0766, IODIDE ION, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Lim, K, Zhang, H, Tempczyk, A, Bonander, N, Toedt, J, Howard, A, Eisenstein, E, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2002-10-02 | | Release date: | 2003-02-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the YibK methyltransferase from Haemophilus influenzae (HI0766): a Cofactor Bound at a Site Formed by a Knot
Proteins, 51, 2003
|
|
1LV3
 
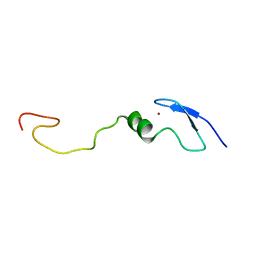 | | Solution NMR Structure of Zinc Finger Protein yacG from Escherichia coli. Northeast Structural Genomics Consortium Target ET92. | | Descriptor: | HYPOTHETICAL PROTEIN YacG, ZINC ION | | Authors: | Ramelot, T.A, Cort, J.R, Yee, A.A, Semesi, A, Edwards, A.M, Arrowsmith, C.H, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-05-24 | | Release date: | 2002-09-11 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the Escherichia coli protein YacG: a novel sequence motif in the zinc-finger family of proteins.
Proteins, 49, 2002
|
|
6R7X
 
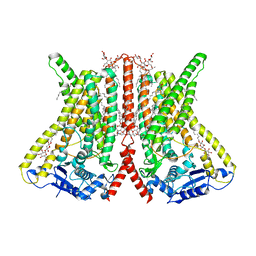 | | CryoEM structure of calcium-bound human TMEM16K / Anoctamin 10 in detergent (2mM Ca2+, closed form) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Anoctamin-10, CALCIUM ION, ... | | Authors: | Pike, A.C.W, Bushell, S.R, Shintre, C.A, Tessitore, A, Baronina, A, Chu, A, Mukhopadhyay, S, Shrestha, L, Chalk, R, Burgess-Brown, N.A, Love, J, Huiskonen, J.T, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-03-29 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | The structural basis of lipid scrambling and inactivation in the endoplasmic reticulum scramblase TMEM16K.
Nat Commun, 10, 2019
|
|
1MW5
 
 | | Structure of HI1480 from Haemophilus influenzae | | Descriptor: | HYPOTHETICAL PROTEIN HI1480 | | Authors: | Lim, K, Sarikaya, E, Howard, A, Galkin, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2002-09-27 | | Release date: | 2003-11-18 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel structure and nucleotide binding properties of HI1480 from Haemophilus influenzae: a protein with no known sequence homologues
PROTEINS: STRUCT.,FUNCT.,GENET., 56, 2004
|
|
6XTJ
 
 | | The high resolution structure of the FERM domain of human FERMT2 | | Descriptor: | CITRIC ACID, Fermitin family homolog 2,Fermitin family homolog 2,Fermitin family homolog 2 | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-01-16 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The high resolution structure of the FERM domain of human FERMT2
To Be Published
|
|
5WOF
 
 | | 1.65 ANGSTROM STRUCTURE OF THE DYNEIN LIGHT CHAIN 1 FROM PLASMODIUM FALCIPARUM | | Descriptor: | Dynein light chain 1, putative | | Authors: | Walker, J.R, Lew, J, Amani, M, Alam, Z, Wasney, G, Boulanger, K, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Hui, R, Botchkarev, A, Vedadi, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-08-02 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Genome-scale Protein Expression and Structural Biology of
Plasmodium Falciparum and Related Apicomplexan Organisms.
MOL.BIOCHEM.PARASITOL., 151, 2007
|
|
6RV3
 
 | | Crystal structure of the human two pore domain potassium ion channel TASK-1 (K2P3.1) in a closed conformation with a bound inhibitor BAY 1000493 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL HEMISUCCINATE, DECYL-BETA-D-MALTOPYRANOSIDE, ... | | Authors: | Rodstrom, K.E.J, Pike, A.C.W, Zhang, W, Quigley, A, Speedman, D, Mukhopadhyay, S.M.M, Shrestha, L, Chalk, R, Venkaya, S, Bushell, S.R, Tessitore, A, Burgess-Brown, N, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-05-30 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A lower X-gate in TASK channels traps inhibitors within the vestibule.
Nature, 582, 2020
|
|
6R7Y
 
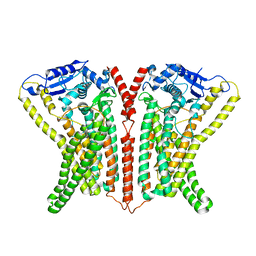 | | CryoEM structure of calcium-bound human TMEM16K / Anoctamin 10 in detergent (low Ca2+, closed form) | | Descriptor: | Anoctamin-10, CALCIUM ION | | Authors: | Pike, A.C.W, Bushell, S.R, Shintre, C.A, Tessitore, A, Chu, A, Mukhopadhyay, S, Shrestha, L, Chalk, R, Burgess-Brown, N.A, Love, J, Huiskonen, J.T, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-03-29 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The structural basis of lipid scrambling and inactivation in the endoplasmic reticulum scramblase TMEM16K.
Nat Commun, 10, 2019
|
|
6R65
 
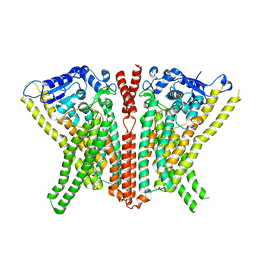 | | Crystal Structure of human TMEM16K / Anoctamin 10 (Form 2) | | Descriptor: | Anoctamin-10, CALCIUM ION | | Authors: | Bushell, S.R, Pike, A.C.W, Chu, A, Tessitore, A, Rotty, B, Mukhopadhyay, S, Kupinska, K, Shrestha, L, Borkowska, O, Chalk, R, Burgess-Brown, N.A, Love, J, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-03-26 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The structural basis of lipid scrambling and inactivation in the endoplasmic reticulum scramblase TMEM16K.
Nat Commun, 10, 2019
|
|
6R7Z
 
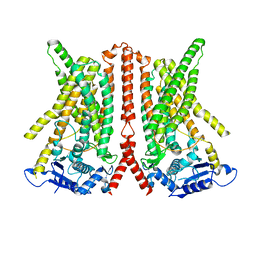 | | CryoEM structure of calcium-free human TMEM16K / Anoctamin 10 in detergent (closed form) | | Descriptor: | Anoctamin-10 | | Authors: | Pike, A.C.W, Bushell, S.R, Shintre, C.A, Tessitore, A, Chu, A, Mukhopadhyay, S, Shrestha, L, Chalk, R, Burgess-Brown, N.A, Love, J, Huiskonen, J.T, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-03-29 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (5.14 Å) | | Cite: | The structural basis of lipid scrambling and inactivation in the endoplasmic reticulum scramblase TMEM16K.
Nat Commun, 10, 2019
|
|
4GD4
 
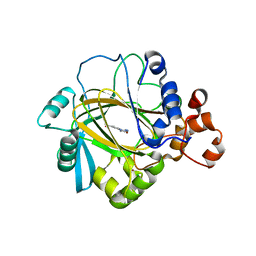 | | Crystal Structure of JMJD2A Complexed with Inhibitor | | Descriptor: | 2-(1H-pyrazol-3-yl)pyridine-4-carboxylic acid, CHLORIDE ION, Lysine-specific demethylase 4A, ... | | Authors: | King, O.N.F, Krojer, T, Arrowsmith, C.H, Edwards, A, Bountra, C, McDonough, M.A, Schofield, C.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-07-31 | | Release date: | 2012-08-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal Structure of JMJD2A Complexed with Inhibitor
TO BE PUBLISHED
|
|
5A3N
 
 | | Crystal structure of human PLU-1 (JARID1B) in complex with KDOAM25a | | Descriptor: | 1,2-ETHANEDIOL, 2-[[[2-[2-(dimethylamino)ethyl-ethyl-amino]-2-oxidanylidene-ethyl]amino]methyl]pyridine-4-carboxamide, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Srikannathasan, V, Johansson, C, Gileadi, C, Nuzzi, A, Ruda, G.F, Kopec, J, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Brennan, P, Oppermann, U. | | Deposit date: | 2015-06-02 | | Release date: | 2015-07-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Potent and Selective KDM5 Inhibitor Stops Cellular Demethylation of H3K4me3 at Transcription Start Sites and Proliferation of MM1S Myeloma Cells.
Cell Chem Biol, 24, 2017
|
|
3DCI
 
 | | The Structure of a putative arylesterase from Agrobacterium tumefaciens str. C58 | | Descriptor: | ACETIC ACID, Arylesterase, CHLORIDE ION, ... | | Authors: | Cuff, M.E, Xu, X, Zheng, H, Binkowski, T.A, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-06-03 | | Release date: | 2008-09-30 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structure of a putative arylesterase from Agrobacterium tumefaciens str. C58
TO BE PUBLISHED
|
|
3DLJ
 
 | | Crystal structure of human carnosine dipeptidase 1 | | Descriptor: | Beta-Ala-His dipeptidase, SULFATE ION, UNKNOWN ATOM OR ION, ... | | Authors: | Dong, A, Dobrovetsky, E, Seitova, A, He, H, Tempel, W, Kozieradzki, I, Arrowsmith, C.H, Weigelt, J, Bountra, C, Edwards, A.M, Bochkarev, A, Cossar, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-06-27 | | Release date: | 2008-08-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure of human carnosine dipeptidase 1.
To be Published
|
|
6EK6
 
 | | Crystal structure of KDM5B in complex with S49195a. | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Lysine-specific demethylase 5B,Lysine-specific demethylase 5B, ... | | Authors: | Srikannathasan, V, Szykowska, A, Newman, J.A, Ruda, G.F, Strain-Damerell, C, Burgess-Brown, N.A, Vazquez-Rodriguez, S, Wright, M, Brennan, P.E, Arrowsmith, C.H, Edwards, A, Bountra, C, Oppermann, U, Huber, K, von Delft, F. | | Deposit date: | 2017-09-25 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of KDM5B in complex with S49195a.
To be published
|
|
5WCF
 
 | | Human HMT1 hnRNP methyltransferase-like protein 6 (S. cerevisiae) | | Descriptor: | (5R)-4-(5-bromofuran-2-carbonyl)-5-(4-fluorophenyl)-7-methyl-1,3,4,5-tetrahydro-2H-1,4-benzodiazepin-2-one, Protein arginine N-methyltransferase 6, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Dong, A, Zeng, H, Hutch, A, Seitova, A, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-06-30 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The Crystal Structure of Human HMT1 hnRNP methyltransferase-like protein 6 in complex with MTLLE1441
to be published
|
|
3CY3
 
 | | Crystal structure of human proto-oncogene serine threonine kinase (PIM1) in complex with a consensus peptide and the JNK inhibitor V | | Descriptor: | (2S)-1,3-benzothiazol-2-yl{2-[(2-pyridin-3-ylethyl)amino]pyrimidin-4-yl}ethanenitrile, 1,2-ETHANEDIOL, Pimtide peptide, ... | | Authors: | Filippakopoulos, P, Bullock, A, Fedorov, O, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-04-25 | | Release date: | 2008-07-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Proto-oncogene serine threonine kinase (PIM1) in complex with a consensus peptide and the JNK inhibitor V.
To be Published
|
|
4URA
 
 | | Crystal structure of human JMJD2A in complex with compound 14a | | Descriptor: | 1,2-ETHANEDIOL, 2-(2H-1,2,3-triazol-4-yl)pyridine-4-carboxylic acid, LYSINE-SPECIFIC DEMETHYLASE 4A, ... | | Authors: | Krojer, T, England, K.S, Vollmar, M, Crawley, L, Williams, E, Riesebos, E, Szykowska, A, Burgess-Brown, N, Oppermann, U, Brennan, P.E, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2014-06-27 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Optimisation of a triazolopyridine based histone demethylase inhibitor yields a potent and selective KDM2A (FBXL11) inhibitor.
Medchemcomm, 5, 2014
|
|
6DVR
 
 | | Crystal structure of human CARM1 with (R)-SKI-72 | | Descriptor: | (2R,5S)-2-amino-6-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]-5-[(benzylamino)methyl]-N-[2-(4-methoxyphenyl)ethyl]hexanamide (non-preferred name), 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, Histone-arginine methyltransferase CARM1, ... | | Authors: | Dong, A, Zeng, H, Hutchinson, A, Seitova, A, Luo, M, Cai, X.C, Ke, W, Wang, J, Shi, C, Zheng, W, Lee, J.P, Ibanez, G, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-06-25 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal structure of human CARM1 with (R)-SKI-72
to be published
|
|
5A7O
 
 | | Crystal structure of human JMJD2A in complex with compound 42 | | Descriptor: | 1,2-ETHANEDIOL, 2-[5-(2-methoxyethanoylamino)-2-oxidanyl-phenyl]pyridine-4-carboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Nowak, R, Velupillai, S, Krojer, T, Gileadi, C, Johansson, C, Korczynska, M, Le, D.D, Younger, N, Gregori-Puigjane, E, Tumber, A, Iwasa, E, Pollock, S.B, Ortiz Torres, I, Pinkas, D.M, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Shoichet, B.K, Fujimori, D.G, Oppermann, U. | | Deposit date: | 2015-07-09 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Docking and Linking of Fragments to Discover Jumonji Histone Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
