1F04
 
 | | SOLUTION STRUCTURE OF OXIDIZED BOVINE MICROSOMAL CYTOCHROME B5 MUTANT (E44A, E48A, E56A, D60A) AND ITS INTERACTION WITH CYTOCHROME C | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wu, Y.B, Lu, J, Qian, C.M, Tang, W.X, Li, E.C, Wang, J.F, Wang, Y.H, Wang, W.H, Lu, J.X, Xie, Y, Huang, Z.X. | | Deposit date: | 2000-05-14 | | Release date: | 2000-06-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of cytochrome b(5) mutant (E44/48/56A/D60A) and its interaction with cytochrome c.
Eur.J.Biochem., 268, 2001
|
|
2AWT
 
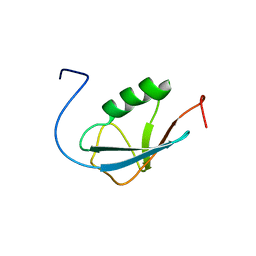 | | Solution Structure of Human Small Ubiquitin-Like Modifier Protein Isoform 2 (SUMO-2) | | Descriptor: | Small ubiquitin-related modifier 2 | | Authors: | Chang, C.K, Wang, Y.H, Chung, T.L, Chang, C.F, Li, S.S.L, Huang, T.H. | | Deposit date: | 2005-09-02 | | Release date: | 2006-10-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Human Small Ubiquitin-Like Modifier Protein Isoform 2 (SUMO-2)
To be Published
|
|
7SPR
 
 | |
8K3G
 
 | |
4MXP
 
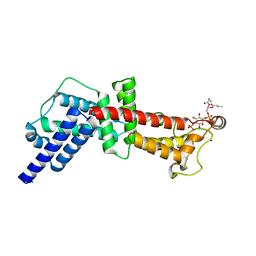 | | Structural Basis for PI(4)P-Specific Membrane Recruitment of the Legionella pneumophila Effector DrrA/SidM | | Descriptor: | (2R)-3-{[(R)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dibutanoate, Defects in Rab1 recruitment protein A, SODIUM ION | | Authors: | Del Campo, C.M, Mishra, A.K, Wang, Y.H, Roy, C.R, Janmey, P.A, Lambright, D.G. | | Deposit date: | 2013-09-26 | | Release date: | 2014-03-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Basis for PI(4)P-Specific Membrane Recruitment of the Legionella pneumophila Effector DrrA/SidM.
Structure, 22, 2014
|
|
6KRJ
 
 | |
6KSR
 
 | |
6LKQ
 
 | | The Structural Basis for Inhibition of Ribosomal Translocation by Viomycin | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Zhang, L, Wang, Y.H, Lancaster, L, Zhou, J, Noller, H.F. | | Deposit date: | 2019-12-20 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structural basis for inhibition of ribosomal translocation by viomycin.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
