7EG7
 
 | | TFIID-based core PIC on SCP promoter | | Descriptor: | DNA (79-MER), DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, ... | | Authors: | Chen, X, Qi, Y, Hou, H, Wang, X, Wu, Z, Li, J, Xu, Y. | | Deposit date: | 2021-03-24 | | Release date: | 2021-05-05 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Structural insights into preinitiation complex assembly on core promoters.
Science, 372, 2021
|
|
7EG9
 
 | | TFIID-based intermediate PIC on SCP promoter | | Descriptor: | DNA (79-MER), DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, ... | | Authors: | Chen, X, Qi, Y, Wang, X, Wu, Z, Li, J, Xu, Y. | | Deposit date: | 2021-03-24 | | Release date: | 2021-05-05 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insights into preinitiation complex assembly on core promoters.
Science, 372, 2021
|
|
7EGA
 
 | | TFIID-based intermediate PIC on PUMA promoter | | Descriptor: | DNA (85-MER), DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, ... | | Authors: | Chen, X, Wu, Z, Hou, H, Qi, Y, Wang, X, Li, J, Xu, Y. | | Deposit date: | 2021-03-24 | | Release date: | 2021-05-05 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insights into preinitiation complex assembly on core promoters.
Science, 372, 2021
|
|
7D4G
 
 | |
2ZNV
 
 | | Crystal structure of human AMSH-LP DUB domain in complex with Lys63-linked ubiquitin dimer | | Descriptor: | 1,2-ETHANEDIOL, AMSH-like protease, Ubiquitin, ... | | Authors: | Sato, Y, Azusa, Y, Yamagata, A, Mimura, H, Wang, X, Yamashita, M, Ookata, K, Nureki, O, Iwai, K, Komada, M, Fukai, S. | | Deposit date: | 2008-05-01 | | Release date: | 2008-09-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains
Nature, 455, 2008
|
|
8TW0
 
 | | Crystal Structure of a synthetic ABC heterotrimeric Collagen-like Peptide at 1.53 A | | Descriptor: | Collagen Mimetic Peptide A, Collagen Mimetic Peptide B, Collagen Mimetic Peptide C, ... | | Authors: | Miller, M.D, Cole, C.C, Xu, W, Walker, D.R, Hulgan, S.A.H, Pogostin, B.H, Swain, J.W.R, Duella, R, Misiura, M, Wang, X, Kolomeisky, A.B, Phillips Jr, G.N, Hartgerink, J.D. | | Deposit date: | 2023-08-18 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Heterotrimeric collagen helix with high specificity of assembly results in a rapid rate of folding.
Nat.Chem., 2024
|
|
2XSP
 
 | | Structure of Cellobiohydrolase 1 (Cel7A) from Heterobasidion annosum | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CELLULOSE 1,4-BETA-CELLOBIOSIDASE, ... | | Authors: | Haddad-momeni, M, Hansson, H, Mikkelsen, N.E, Wang, X, Svedberg, J, Sandgren, M, Stahlberg, J. | | Deposit date: | 2010-09-29 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural, Biochemical, and Computational Characterization of the Glycoside Hydrolase Family 7 Cellobiohydrolase of the Tree-Killing Fungus Heterobasidion Irregulare.
J.Biol.Chem., 288, 2013
|
|
2ZNR
 
 | | Crystal structure of the DUB domain of human AMSH-LP | | Descriptor: | 1,2-ETHANEDIOL, AMSH-like protease, PRASEODYMIUM ION, ... | | Authors: | Sato, Y, Azusa, Y, Yamagata, A, Mimura, H, Wang, X, Yamashita, M, Ookata, K, Nureki, O, Iwai, K, Komada, M, Fukai, S. | | Deposit date: | 2008-05-01 | | Release date: | 2008-09-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains
Nature, 455, 2008
|
|
2KS6
 
 | |
7WPO
 
 | | Structure of NeoCOV RBD binding to Bat37 ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cao, L, Wang, X, Tortorici, M.A, Veesler, D. | | Deposit date: | 2022-01-24 | | Release date: | 2022-11-30 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Close relatives of MERS-CoV in bats use ACE2 as their functional receptors.
Nature, 612, 2022
|
|
7WPZ
 
 | | Structure of PDF-2180-COV RBD binding to Bat37 ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Cao, L, Wang, X, Tortorici, M.A, Veesler, D. | | Deposit date: | 2022-01-24 | | Release date: | 2022-11-30 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Close relatives of MERS-CoV in bats use ACE2 as their functional receptors.
Nature, 612, 2022
|
|
7X25
 
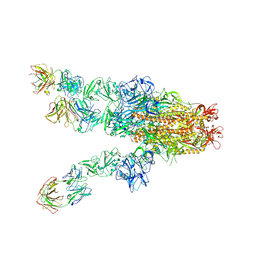 | | MERS-CoV spike complex with S41 neutralizing antibody Fab Class4 (2u1d RBD with 3Fab) | | Descriptor: | Spike glycoprotein, antibody S41 heavy chain, antibody S41 light chain | | Authors: | Zeng, J, Zhang, S, Zhou, H, Wang, X. | | Deposit date: | 2022-02-25 | | Release date: | 2023-01-18 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Cryoelectron microscopy structures of a human neutralizing antibody bound to MERS-CoV spike glycoprotein.
Front Microbiol, 13, 2022
|
|
2RO1
 
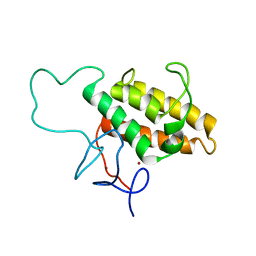 | | NMR Solution Structures of Human KAP1 PHD finger-bromodomain | | Descriptor: | Transcription intermediary factor 1-beta, ZINC ION | | Authors: | Zeng, L, Yap, K.L, Ivanov, A.V, Wang, X, Mujtaba, S, Plotnikova, O, Rauscher, F.J. | | Deposit date: | 2008-03-04 | | Release date: | 2008-05-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural insights into human KAP1 PHD finger-bromodomain and its role in gene silencing
Nat.Struct.Mol.Biol., 15, 2008
|
|
2JXL
 
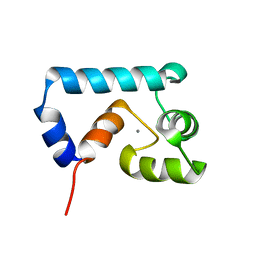 | | Solution structure of cardiac N-domain troponin C mutant F77W-V82A | | Descriptor: | CALCIUM ION, Troponin C, slow skeletal and cardiac muscles | | Authors: | Julien, O, Sun, Y, Wang, X, Lindhout, D.A, Thiessen, A, Irving, M, Sykes, B.D. | | Deposit date: | 2007-11-20 | | Release date: | 2007-12-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Tryptophan Mutants of Cardiac Troponin C: 3D Structure, Troponin I Affinity, and in Situ Activity.
Biochemistry, 47, 2008
|
|
2N62
 
 | | ddFLN5+110 | | Descriptor: | gelation factor, secretion monitor chimera | | Authors: | Cabrita, L.D, Cassaignau, A.M.E, Launay, H.M.M, Waudby, C.A, Camilloni, C, Robertson, A.L, Wang, X, Wlodarski, T, Wentink, A.S, Vendruscolo, M, Dobson, C.M, Christodoulou, J. | | Deposit date: | 2015-08-10 | | Release date: | 2016-03-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A structural ensemble of a ribosome-nascent chain complex during cotranslational protein folding.
Nat.Struct.Mol.Biol., 23, 2016
|
|
2PQ6
 
 | |
2N6F
 
 | | Structure of Pleiotrophin | | Descriptor: | Pleiotrophin | | Authors: | Ryan, E.O, Shen, D, Wang, X. | | Deposit date: | 2015-08-20 | | Release date: | 2016-04-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural studies reveal an important role for the pleiotrophin C-terminus in mediating interactions with chondroitin sulfate.
Febs J., 283, 2016
|
|
2KCK
 
 | | NMR solution structure of the Northeast Structural Genomics Consortium (NESG) target MrR121A | | Descriptor: | TPR repeat | | Authors: | Barb, A.W, Lee, H.-W, Wang, X, Lee, D, Jiang, M, Ciccosanti, C, Xiao, R, Nair, R, Everett, J.K, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Prestegard, J.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-22 | | Release date: | 2009-01-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the Northeast Structural Genomics Target MrR121A
To be Published
|
|
7F7E
 
 | | SARS-CoV-2 S protein RBD in complex with A5-10 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of A5-10 Fab, Light chain of A5-10 Fab, ... | | Authors: | Dou, Y, Wang, X, Wang, K, Liu, P, Lu, B. | | Deposit date: | 2021-06-29 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Etesevimab in combination with JS026 neutralizing SARS-CoV-2 and its variants.
Emerg Microbes Infect, 11, 2022
|
|
8J6W
 
 | |
8OR1
 
 | | Co-crystal strucutre of PD-L1 with low molecular weight inhibitor | | Descriptor: | 5-[[5-[[2-chloranyl-3-(2-fluorophenyl)phenyl]methoxy]-2-[(~{E})-2-hydroxyethyliminomethyl]phenoxy]methyl]pyridine-3-carbonitrile, Programmed cell death 1 ligand 1 | | Authors: | Zhang, H, Zhou, S, Wu, C, Zhu, M, Yu, Q, Wang, X, Awadasseid, A, Plewka, J, Magiera-Mularz, K, Wu, Y, Zhang, W. | | Deposit date: | 2023-04-12 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Design, Synthesis, and Antitumor Activity Evaluation of 2-Arylmethoxy-4-(2,2'-dihalogen-substituted biphenyl-3-ylmethoxy) Benzylamine Derivatives as Potent PD-1/PD-L1 Inhibitors.
J.Med.Chem., 66, 2023
|
|
7VRV
 
 | | VAS5 Spike (1 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhen, C, Wang, X. | | Deposit date: | 2021-10-25 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | VAS5 Spike(1 RBD up)
To Be Published
|
|
6JHS
 
 | | The cryo-EM structure of HAV bound to a neutralizing antibody-F7 | | Descriptor: | FAB Heavy Chain, FAB Light Chain, VP1, ... | | Authors: | Cao, L, Liu, P, Yang, P, Gao, Q, Li, H, Sun, Y, Zhu, L, Lin, J, Su, D, Rao, Z, Wang, X. | | Deposit date: | 2019-02-19 | | Release date: | 2020-03-18 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural basis for neutralization of hepatitis A virus informs a rational design of highly potent inhibitors.
Plos Biol., 17, 2019
|
|
6KG3
 
 | |
6KH2
 
 | |
