5BTB
 
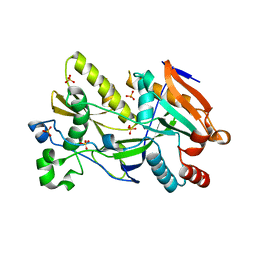 | | Crystal Structure of Ashbya gossypii Rai1 | | Descriptor: | AFR263Cp, SULFATE ION | | Authors: | Wang, V.Y, Tong, L. | | Deposit date: | 2015-06-02 | | Release date: | 2015-07-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and biochemical studies of the distinct activity profiles of Rai1 enzymes.
Nucleic Acids Res., 43, 2015
|
|
5BTE
 
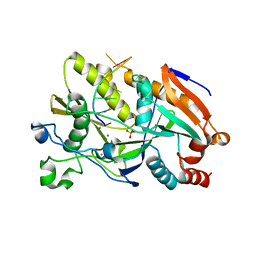 | |
5BTH
 
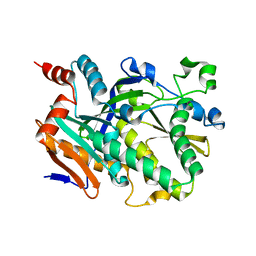 | | Crystal structure of Candida albicans Rai1 | | Descriptor: | Decapping nuclease RAI1 | | Authors: | Wang, V.Y, Tong, L. | | Deposit date: | 2015-06-03 | | Release date: | 2015-07-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and biochemical studies of the distinct activity profiles of Rai1 enzymes.
Nucleic Acids Res., 43, 2015
|
|
5BUD
 
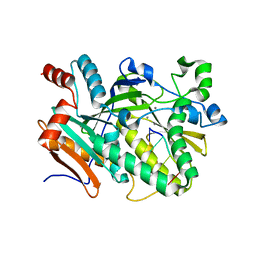 | |
5BTO
 
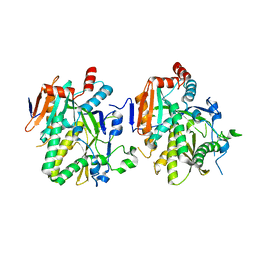 | |
6DKN
 
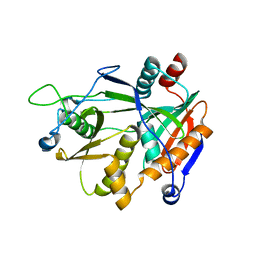 | |
7VUP
 
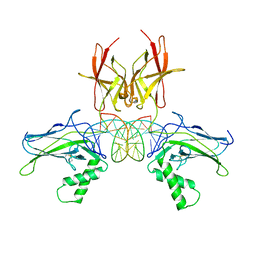 | |
7VUQ
 
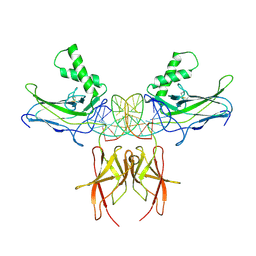 | |
7W7L
 
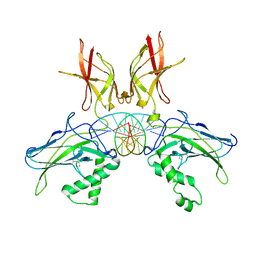 | |
2V2T
 
 | | X-ray structure of a NF-kB p50-RelB-DNA complex | | Descriptor: | 5'-D(*CP*GP*GP*GP*AP*AP*TP*TP*CP*CP*CP)-3', NUCLEAR FACTOR NF-KAPPA-B P105 SUBUNIT, TRANSCRIPTION FACTOR RELB | | Authors: | Moorthy, A.K, Huang, D.B, Wang, V.Y, Vu, D, Ghosh, G. | | Deposit date: | 2007-06-07 | | Release date: | 2007-07-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | X-Ray Structure of a NF-kappaB p50/Relb/DNA Complex Reveals Assembly of Multiple Dimers on Tandem kappaB Sites.
J.Mol.Biol., 373, 2007
|
|
9BDZ
 
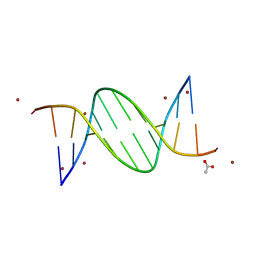 | | TA-centric NF-kappaB RelA binding DNA | | Descriptor: | ACETATE ION, ZINC ION, double-stranded DNA | | Authors: | Biswas, T, Shahabi, S, Tsodikov, O.V, Wang, V, Ghosh, G. | | Deposit date: | 2024-04-13 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Transient interactions modulate the affinity of NF-kappa B transcription factors for DNA.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9BE1
 
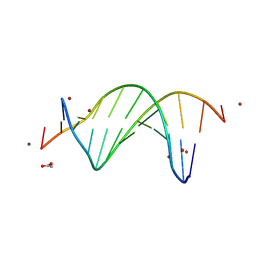 | | CG-centric NF-kappaB RelA binding DNA | | Descriptor: | ACETATE ION, DNA (5'-D(*CP*GP*GP*GP*AP*AP*TP*TP*TP*CP*CP*GP*CP*GP*GP*AP*AP*AP*TP*TP*CP*CP*CP*G)-3'), ZINC ION | | Authors: | Biswas, T, Shahabi, S, Tsodikov, O.V, Wang, V, Ghosh, G. | | Deposit date: | 2024-04-13 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Transient interactions modulate the affinity of NF-kappa B transcription factors for DNA.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
3OR1
 
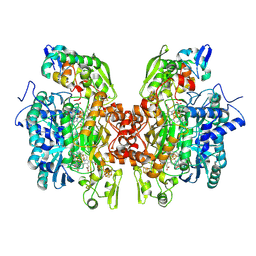 | | Crystal structure of dissimilatory sulfite reductase I (DsrI) | | Descriptor: | IRON/SULFUR CLUSTER, SIROHEME, SULFITE ION, ... | | Authors: | Hsieh, Y.C, Liu, M.Y, Wang, V.C.C, Chiang, Y.L, Liu, E.H, Wu, W.G, Chan, S.I, Chen, C.J. | | Deposit date: | 2010-09-06 | | Release date: | 2010-09-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structure insights into the enzyme catalysis from comparison of three forms of dissimilatory sulfite reductase from Desulfovibrio gigas
To be Published
|
|
3OR2
 
 | | Crystal structure of dissimilatory sulfite reductase II (DsrII) | | Descriptor: | IRON/SULFUR CLUSTER, SIROHEME, SULFITE ION, ... | | Authors: | Hsieh, Y.C, Liu, M.Y, Wang, V.C.C, Chiang, Y.L, Liu, E.H, Wu, W.G, Chan, S.I, Chen, C.J. | | Deposit date: | 2010-09-06 | | Release date: | 2010-09-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Dissimilatory Sulfite Reductase, Sulfate Reduction
To be Published
|
|
7CLI
 
 | | Structure of NF-kB p52 homodimer bound to P-Selectin kB DNA fragment | | Descriptor: | DNA (5'-D(*CP*AP*AP*GP*GP*GP*GP*TP*CP*AP*CP*CP*CP*CP*CP*TP*TP*C)-3'), DNA (5'-D(*GP*AP*AP*GP*GP*GP*GP*GP*TP*GP*AP*CP*CP*CP*CP*TP*TP*G)-3'), Nuclear factor NF-kappa-B p52 subunit | | Authors: | Meshcheryakov, V.A, Wang, V.Y.-F. | | Deposit date: | 2020-07-21 | | Release date: | 2021-07-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of NF-kappa B p52 homodimer-DNA complexes rationalize binding mechanisms and transcription activation.
Elife, 12, 2023
|
|
