2MM5
 
 | |
2MAU
 
 | |
2LYI
 
 | |
2PMU
 
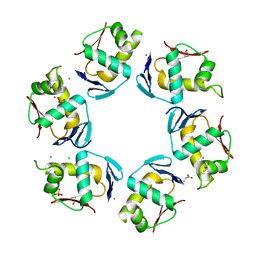 | | Crystal structure of the DNA-binding domain of PhoP | | Descriptor: | CHLORIDE ION, GLYCINE, PHOSPHATE ION, ... | | Authors: | Wang, S. | | Deposit date: | 2007-04-23 | | Release date: | 2008-02-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.779 Å) | | Cite: | Structure of the DNA-binding domain of the response regulator PhoP from Mycobacterium tuberculosis
Biochemistry, 46, 2007
|
|
1D1P
 
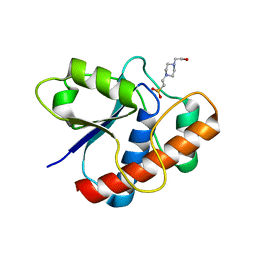 | | CRYSTAL STRUCTURE OF A YEAST LOW MOLECULAR WEIGHT PROTEIN TYROSINE PHOSPHATASE (LTP1) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, TYROSINE PHOSPHATASE | | Authors: | Wang, S, Tabernero, L, Zhang, M, Harms, E, Van Etten, R.L, Stauffacher, C.V. | | Deposit date: | 1999-09-20 | | Release date: | 2000-03-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of a low-molecular weight protein tyrosine phosphatase from Saccharomyces cerevisiae and its complex with the substrate p-nitrophenyl phosphate.
Biochemistry, 39, 2000
|
|
1D1Q
 
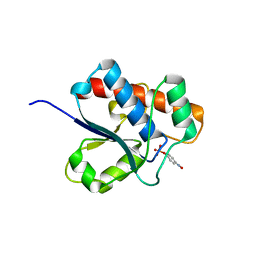 | | CRYSTAL STRUCTURE OF A YEAST LOW MOLECULAR WEIGHT PROTEIN TYROSINE PHOSPHATASE (LTP1) COMPLEXED WITH THE SUBSTRATE PNPP | | Descriptor: | 4-NITROPHENYL PHOSPHATE, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Wang, S, Tabernero, L, Zhang, M, Harms, E, Van Etten, R.L, Staufacher, C.V. | | Deposit date: | 1999-09-20 | | Release date: | 2000-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of a low-molecular weight protein tyrosine phosphatase from Saccharomyces cerevisiae and its complex with the substrate p-nitrophenyl phosphate.
Biochemistry, 39, 2000
|
|
6AKR
 
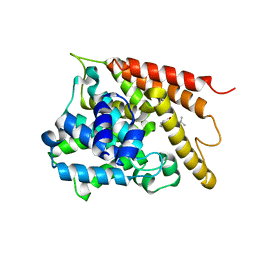 | | Crystal structure of the PDE4D catalytic domain in complex with osthole | | Descriptor: | 7-methoxy-8-(3-methylbut-2-enyl)chromen-2-one, ZINC ION, cAMP-specific 3',5'-cyclic phosphodiesterase 4D | | Authors: | Wang, S, Huo, Y.W, Xie, Y. | | Deposit date: | 2018-09-03 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.326 Å) | | Cite: | Airway relaxation mechanisms and structural basis of osthole for improving lung function in asthma.
Sci.Signal., 13, 2020
|
|
7DBP
 
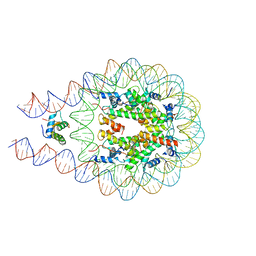 | | Linker histone defines structure and self-association behaviour of the 177 bp human chromosome | | Descriptor: | DNA (175-MER), Histone H1.0, Histone H2A type 1-B/E, ... | | Authors: | Wang, S, Vogirala, K.V, Soman, A, Liu, Z.B. | | Deposit date: | 2020-10-21 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Linker histone defines structure and self-association behaviour of the 177 bp human chromatosome.
Sci Rep, 11, 2021
|
|
2A86
 
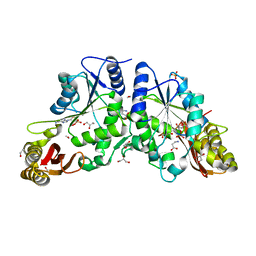 | | Crystal structure of A Pantothenate synthetase complexed with AMP and beta-alanine | | Descriptor: | ADENOSINE MONOPHOSPHATE, BETA-ALANINE, ETHANOL, ... | | Authors: | Wang, S, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2005-07-07 | | Release date: | 2006-02-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of the Pantothenate Synthetase from Mycobacterium tuberculosis, Snapshots of the Enzyme in Action.
Biochemistry, 45, 2006
|
|
2A84
 
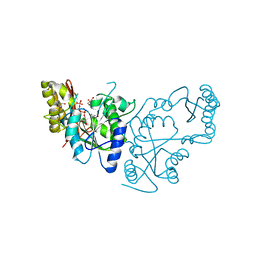 | | Crystal structure of A Pantothenate synthetase complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Wang, S, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2005-07-07 | | Release date: | 2006-02-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of the Pantothenate Synthetase from Mycobacterium tuberculosis, Snapshots of the Enzyme in Action.
Biochemistry, 45, 2006
|
|
2A88
 
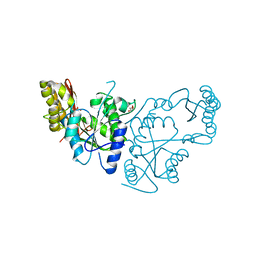 | | Crystal structure of A Pantothenate synthetase, apo enzyme in C2 space group | | Descriptor: | GLYCEROL, Pantoate--beta-alanine ligase, SULFATE ION | | Authors: | Wang, S, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2005-07-07 | | Release date: | 2006-02-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the Pantothenate Synthetase from Mycobacterium tuberculosis, Snapshots of the Enzyme in Action.
Biochemistry, 45, 2006
|
|
2A7X
 
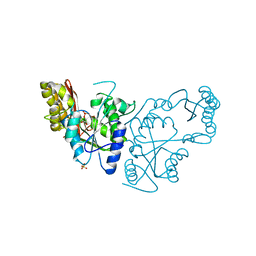 | | Crystal Structure of A Pantothenate synthetase complexed with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCEROL, Pantoate-beta-alanine ligase, ... | | Authors: | Wang, S, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2005-07-06 | | Release date: | 2006-02-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the Pantothenate Synthetase from Mycobacterium tuberculosis, Snapshots of the Enzyme in Action.
Biochemistry, 45, 2006
|
|
2I8E
 
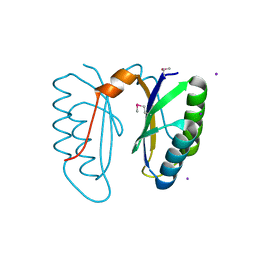 | | Structure of SSO1404, a predicted DNA repair-associated protein from Sulfolobus solfataricus P2 | | Descriptor: | Hypothetical protein, IODIDE ION | | Authors: | Wang, S, Zimmerman, M.D, Kudritska, M, Chruszcz, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-09-01 | | Release date: | 2006-09-26 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | A novel family of sequence-specific endoribonucleases associated with the clustered regularly interspaced short palindromic repeats.
J.Biol.Chem., 283, 2008
|
|
2XMY
 
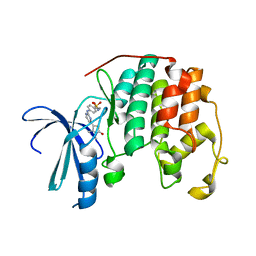 | | Discovery and Characterisation of 2-Anilino-4-(thiazol-5-yl) pyrimidine Transcriptional CDK Inhibitors as Anticancer Agents | | Descriptor: | 4-[4-(3,4-DIMETHYL-2-OXO-2,3-DIHYDRO-THIAZOL-5-YL)-PYRIMIDIN-2-YLAMINO]-N-(2-METHOXY-ETHYL)-BENZENESULFONAMIDE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Wang, S, Griffiths, G, Midgley, C.A, Barnett, A.L, Cooper, M, Grabarek, J, Ingram, L, Jackson, W, Kontopidis, G, McClue, S.J, McInnes, C, McLachlan, J, Meades, C, Mezna, M, Stuart, I, Thomas, M.P, Zheleva, D.I, Lane, D.P, Jackson, R.C, Glover, D.M, Blake, D.G, Fischer, P.M. | | Deposit date: | 2010-07-29 | | Release date: | 2010-11-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and Characterisation of 2-Anilino-4-(Thiazol-5-Yl)Pyrimidine Transcriptional Cdk Inhibitors as Anticancer Agents
Chem.Biol., 17, 2010
|
|
2XNB
 
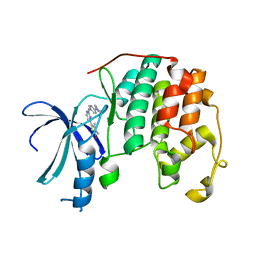 | | Discovery and Characterisation of 2-Anilino-4-(thiazol-5-yl) pyrimidine Transcriptional CDK Inhibitors as Anticancer Agents | | Descriptor: | 3,4-DIMETHYL-5-(2-{[(1Z)-4-PIPERAZIN-1-YLCYCLOHEXA-2,4-DIEN-1-YLIDENE]AMINO}PYRIMIDIN-4-YL)-1,3-THIAZOL-2(3H)-ONE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Wang, S, Griffiths, G, Midgley, C.A, Barnett, A.L, Cooper, M, Grabarek, J, Ingram, L, Jackson, W, Kontopidis, G, McClue, S.J, McInnes, C, McLachlan, J, Meades, C, Mezna, M, Stuart, I, Thomas, M.P, Zheleva, D.I, Lane, D.P, Jackson, R.C, Glover, D.M, Blake, D.G, Fischer, P.M. | | Deposit date: | 2010-08-01 | | Release date: | 2010-11-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery and Characterisation of 2-Anilino-4-(Thiazol-5-Yl)Pyrimidine Transcriptional Cdk Inhibitors as Anticancer Agents
Chem.Biol., 17, 2010
|
|
2FDO
 
 | | Crystal Structure of the Conserved Protein of Unknown Function AF2331 from Archaeoglobus fulgidus DSM 4304 Reveals a New Type of Alpha/Beta Fold | | Descriptor: | Hypothetical protein AF2331 | | Authors: | Wang, S, Kirillova, O, Chruszcz, M, Cymborowski, M.T, Skarina, T, Gorodichtchenskaia, E, Savchenko, A, Edwards, A.M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-14 | | Release date: | 2006-01-31 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of the AF2331 protein from Archaeoglobus fulgidus DSM 4304 forms an unusual interdigitated dimer with a new type of alpha + beta fold.
Protein Sci., 18, 2009
|
|
7XUG
 
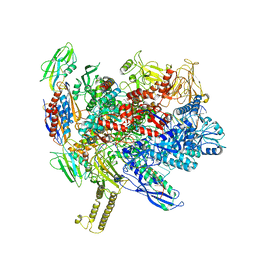 | |
7XUI
 
 | |
2L0O
 
 | | DsbB3 peptide structure in 100% TFE | | Descriptor: | Oxidoreductase that catalyzes reoxidation of DsbA protein disulfide isomerase I | | Authors: | Hwang, S, Hilty, C. | | Deposit date: | 2010-07-08 | | Release date: | 2011-03-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Folding determinants of disulfide bond forming protein B explored by solution nuclear magnetic resonance spectroscopy.
Proteins, 79, 2011
|
|
2L0L
 
 | | DsbB2 peptide structure in 70% TFE | | Descriptor: | Oxidoreductase that catalyzes reoxidation of DsbA protein disulfide isomerase I | | Authors: | Hwang, S, Hilty, C. | | Deposit date: | 2010-07-08 | | Release date: | 2011-03-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Folding determinants of disulfide bond forming protein B explored by solution nuclear magnetic resonance spectroscopy.
Proteins, 79, 2011
|
|
2L0M
 
 | | DsbB2 peptide structure in 100% TFE | | Descriptor: | Oxidoreductase that catalyzes reoxidation of DsbA protein disulfide isomerase I | | Authors: | Hwang, S, Hilty, C. | | Deposit date: | 2010-07-08 | | Release date: | 2011-03-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Folding determinants of disulfide bond forming protein B explored by solution nuclear magnetic resonance spectroscopy.
Proteins, 79, 2011
|
|
2L0N
 
 | | DsbB3 peptide structure in 70% TFE | | Descriptor: | Oxidoreductase that catalyzes reoxidation of DsbA protein disulfide isomerase I | | Authors: | Hwang, S, Hilty, C. | | Deposit date: | 2010-07-08 | | Release date: | 2011-03-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Folding determinants of disulfide bond forming protein B explored by solution nuclear magnetic resonance spectroscopy.
Proteins, 79, 2011
|
|
1GYY
 
 | | The Crystal Structure of YdcE, a 4-Oxalocrotonate Tautomerase Homologue from Escherichia coli, Confirms the Structural Basis for Oligomer Diversity | | Descriptor: | 2-FLUORO-3-(4-HYDROXYPHENYL)-2E-PROPENEOATE, HYPOTHETICAL PROTEIN YDCE | | Authors: | Almrud, J, Kern, A, Wang, S, Czerwinski, R, Johnson, W, Murzin, A, Hackert, M, Whitman, C. | | Deposit date: | 2002-04-30 | | Release date: | 2002-10-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The Crystal Structure of Ydce, a 4-Oxalocrotonate Tautomerase Homologue from Escherichia Coli, Confirms the Structural Basis for Oligomer Diversity
Biochemistry, 41, 2002
|
|
1GYJ
 
 | | The Crystal Structure of YdcE, a 4-Oxalocrotonate Tautomerase Homologue from Escherichia coli, Confirms the Structural Basis for Oligomer Diversity | | Descriptor: | HYPOTHETICAL PROTEIN YDCE | | Authors: | Almrud, J, Kern, A, Wang, S, Czerwinski, R, Johnson, W, Murzin, A, Hackert, M, Whitman, C. | | Deposit date: | 2002-04-23 | | Release date: | 2002-10-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of Ydce, a 4-Oxalocrotonate Tautomerase Homologue from Escherichia Coli, Confirms the Structural Basis for Oligomer Diversity
Biochemistry, 41, 2002
|
|
2AV9
 
 | | Crystal Structure of the PA5185 protein from Pseudomonas Aeruginosa Strain PAO1. | | Descriptor: | SULFATE ION, Thioesterase | | Authors: | Chruszcz, M, Wang, S, Cymborowski, M, Kudritska, M, Evdokimova, E, Edwards, A, Savchenko, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-08-29 | | Release date: | 2005-10-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Function-biased choice of additives for optimization of protein crystallization - the case of the putative thioesterase PA5185 from Pseudomonas aeruginosa PAO1.
Cryst.Growth Des., 8, 2008
|
|
