6CCP
 
 | |
1XK3
 
 | | NADPH- and Ascorbate-Supported Heme Oxygenase Reactions are Distinct. Regiospecificity of Heme Cleavage by the R183E Mutant | | Descriptor: | Heme oxygenase 1, NITRIC OXIDE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wang, J, Lad, L, Poulos, T.L, Ortiz de montellano, P.R. | | Deposit date: | 2004-09-26 | | Release date: | 2004-11-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Regiospecificity determinants of human heme oxygenase: differential NADPH- and ascorbate-dependent heme cleavage by the R183E mutant.
J.Biol.Chem., 280, 2005
|
|
1XK2
 
 | | NADPH- and Ascorbate-Supported Heme Oxygenase Reactions are Distinct. Regiospecificity of Heme Cleavage by the R183E Mutant | | Descriptor: | Heme oxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wang, J, Lad, L, Poulos, T.L, Ortiz de montellano, P.R. | | Deposit date: | 2004-09-26 | | Release date: | 2004-11-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Regiospecificity determinants of human heme oxygenase: differential NADPH- and ascorbate-dependent heme cleavage by the R183E mutant.
J.Biol.Chem., 280, 2005
|
|
5KQL
 
 | |
5KQP
 
 | | Crystal structure of Apo-form LMW-PTP | | Descriptor: | Low molecular weight phosphotyrosine protein phosphatase | | Authors: | Wang, J, Zhang, Z.-Y, Yu, Z.-H. | | Deposit date: | 2016-07-06 | | Release date: | 2016-10-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | Inhibition of low molecular weight protein tyrosine phosphatase by an induced-fit mechanism.
J.Med.Chem., 2016
|
|
7WSB
 
 | | The ternary complex structure of FtmOx1 with a-ketoglutarate and 13-oxo-fumitremorgin B | | Descriptor: | 13-Oxofumitremorgin B, 2-OXOGLUTARIC ACID, COBALT (II) ION, ... | | Authors: | Wang, J, Wang, X.Y, Wang, Y.Y, Yan, W.P. | | Deposit date: | 2022-01-28 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Dissecting the Mechanism of the Nonheme Iron Endoperoxidase FtmOx1 Using Substrate Analogues.
Jacs Au, 2, 2022
|
|
5KQG
 
 | |
5KQM
 
 | | Co-crystal structure of LMW-PTP in complex with MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Low molecular weight phosphotyrosine protein phosphatase | | Authors: | Wang, J, Zhang, Z.-Y, Yu, Z.-H. | | Deposit date: | 2016-07-06 | | Release date: | 2016-10-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Inhibition of low molecular weight protein tyrosine phosphatase by an induced-fit mechanism.
J.Med.Chem., 2016
|
|
1ZA1
 
 | | Structure of wild-type E. coli Aspartate Transcarbamoylase in the presence of CTP at 2.20 A resolution | | Descriptor: | Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Wang, J, Stieglitz, K.A, Cardia, J.P, Kantrowitz, E.R. | | Deposit date: | 2005-04-05 | | Release date: | 2005-06-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for ordered substrate binding and cooperativity in aspartate transcarbamoylase
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1Y7A
 
 | | Structure of D153H/K328W E. coli alkaline phosphatase in presence of cobalt at 1.77 A resolution | | Descriptor: | Alkaline phosphatase, COBALT (II) ION, PHOSPHATE ION, ... | | Authors: | Wang, J, Stieglitz, K, Kantrowitz, E.R. | | Deposit date: | 2004-12-08 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Metal Specificity Is Correlated with Two Crucial Active Site Residues in Escherichia coli Alkaline Phosphatase(,).
Biochemistry, 44, 2005
|
|
6M0A
 
 | | The heme-bound structure of the chloroplast protein At3g03890 | | Descriptor: | AT3G03890 protein, AZIDE ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wang, J, Liu, L. | | Deposit date: | 2020-02-20 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Arabidopsis locus AT3G03890 encodes a dimeric beta-barrel protein implicated in heme degradation.
Biochem.J., 2020
|
|
1Y6V
 
 | | Structure of E. coli Alkaline Phosphatase in presence of cobalt at 1.60 A resolution | | Descriptor: | Alkaline phosphatase, COBALT (II) ION, PHOSPHATE ION, ... | | Authors: | Wang, J, Stieglitz, K, Kantrowitz, E.R. | | Deposit date: | 2004-12-07 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Metal Specificity Is Correlated with Two Crucial Active Site Residues in Escherichia coli Alkaline Phosphatase(,).
Biochemistry, 44, 2005
|
|
1ZA2
 
 | | Structure of wild-type E. coli Aspartate Transcarbamoylase in the presence of CTP, carbamoyl phosphate at 2.50 A resolution | | Descriptor: | Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Wang, J, Stieglitz, K.A, Cardia, J.P, Kantrowitz, E.R. | | Deposit date: | 2005-04-05 | | Release date: | 2005-06-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for ordered substrate binding and cooperativity in aspartate transcarbamoylase
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1ZET
 
 | |
4F7C
 
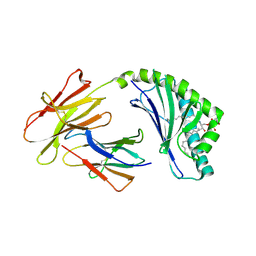 | | Crystal structure of bovine CD1d with bound C12-di-sulfatide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, CD1D antigen, ... | | Authors: | Wang, J, Zajonc, D.M. | | Deposit date: | 2012-05-15 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.864 Å) | | Cite: | Crystal Structures of Bovine CD1d Reveal Altered αGalCer Presentation and a Restricted A' Pocket Unable to Bind Long-Chain Glycolipids.
Plos One, 7, 2012
|
|
4F7E
 
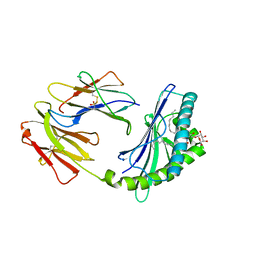 | | Crystal structure of bovine CD1d with bound C16:0-alpha-galactosyl ceramide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, ... | | Authors: | Wang, J, Zajonc, D.M. | | Deposit date: | 2012-05-15 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Bovine CD1d Reveal Altered αGalCer Presentation and a Restricted A' Pocket Unable to Bind Long-Chain Glycolipids.
Plos One, 7, 2012
|
|
5YNW
 
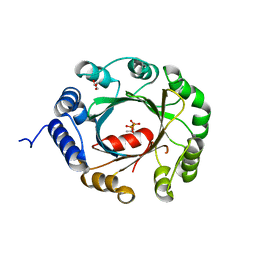 | | Crystal structure of an aromatic prenyltransferase FAMD1 from Fischerella ambigua UTEX 1903 in complex with DMASPP and INN | | Descriptor: | 3-[(Z)-2-isocyanoethenyl]-1H-indole, DIMETHYLALLYL S-THIOLODIPHOSPHATE, GLYCEROL, ... | | Authors: | Wang, J, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-10-25 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural insight into a novel indole prenyltransferase in hapalindole-type alkaloid biosynthesis.
Biochem. Biophys. Res. Commun., 495, 2018
|
|
5YL6
 
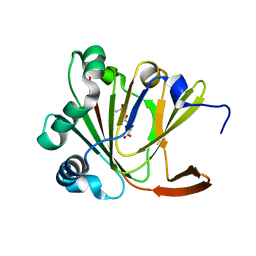 | | Crystal structure of Ofd2 in complex with 2OG from Schizosaccharomyces pombe | | Descriptor: | 2-OXOGLUTARIC ACID, ACETATE ION, FE (III) ION, ... | | Authors: | Wang, J, Gao, Y.X, Guo, G.R, Zhu, Z.L, Teng, M.K, Niu, L.W. | | Deposit date: | 2017-10-17 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Ofd2 in complex with 2OG from Schizosaccharomyces pombe
to be published
|
|
5YLB
 
 | | Crystal structure of Ofd2 from Schizosaccharomyces pombe at 1.80 A | | Descriptor: | FE (III) ION, Uncharacterized protein P8A3.02c | | Authors: | Wang, J, Gao, Y.X, Guo, G.R, Zhu, Z.L, Teng, M.K, Niu, L.W. | | Deposit date: | 2017-10-17 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of Ofd2 from Schizosaccharomyces pombe at 1.80 A
to be published
|
|
5YNT
 
 | | Crystal structure of an aromatic prenyltransferase FAMD1 from Fischerella ambigua UTEX 1903 | | Descriptor: | DIMETHYLALLYL DIPHOSPHATE, GLYCEROL, IMIDAZOLE, ... | | Authors: | Wang, J, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-10-25 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural insight into a novel indole prenyltransferase in hapalindole-type alkaloid biosynthesis.
Biochem. Biophys. Res. Commun., 495, 2018
|
|
5YNU
 
 | | Crystal structure of an aromatic prenyltransferase FAMD1 from Fischerella ambigua UTEX 1903 in complex with INN | | Descriptor: | 3-[(Z)-2-isocyanoethenyl]-1H-indole, DIMETHYLALLYL DIPHOSPHATE, GLYCEROL, ... | | Authors: | Wang, J, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-10-25 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insight into a novel indole prenyltransferase in hapalindole-type alkaloid biosynthesis.
Biochem. Biophys. Res. Commun., 495, 2018
|
|
5YNV
 
 | | Crystal structure of an aromatic prenyltransferase FAMD1 from Fischerella ambigua UTEX 1903 in complex with DMASPP | | Descriptor: | DIMETHYLALLYL S-THIOLODIPHOSPHATE, GLYCEROL, aromatic prenyltransferase | | Authors: | Wang, J, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-10-25 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insight into a novel indole prenyltransferase in hapalindole-type alkaloid biosynthesis.
Biochem. Biophys. Res. Commun., 495, 2018
|
|
7SJR
 
 | | Cryo-EM structure of AdnA-AdnB(W325A) in complex with DNA and AMPPNP | | Descriptor: | DNA (70-MER), DNA helicase, IRON/SULFUR CLUSTER, ... | | Authors: | Wang, J, Warren, G.M, Shuman, S, Patel, D.J. | | Deposit date: | 2021-10-18 | | Release date: | 2021-12-22 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure-activity relationships at a nucleobase-stacking tryptophan required for chemomechanical coupling in the DNA resecting motor-nuclease AdnAB.
Nucleic Acids Res., 50, 2022
|
|
5Z11
 
 | | Crystal Structure of Grass Carp CD8 alpha alpha Homodimers | | Descriptor: | CD8 alpha chain | | Authors: | Wang, J. | | Deposit date: | 2017-12-22 | | Release date: | 2018-04-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of the Grass Carp CD8 alpha
alpha Homodimers Indicates a Dramatic Evolution of CD8 from Ectotherms to Endotherms.
To Be Published
|
|
3HYM
 
 | | Insights into Anaphase Promoting Complex TPR subdomain assembly from a CDC26-APC6 structure | | Descriptor: | Anaphase-promoting complex subunit CDC26, Cell division cycle protein 16 homolog | | Authors: | Wang, J, Dye, B.T, Rajashankar, K.R, Kurinov, I, Schulman, B.A. | | Deposit date: | 2009-06-22 | | Release date: | 2009-08-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Insights into anaphase promoting complex TPR subdomain assembly from a CDC26-APC6 structure.
Nat.Struct.Mol.Biol., 16, 2009
|
|
