6TYD
 
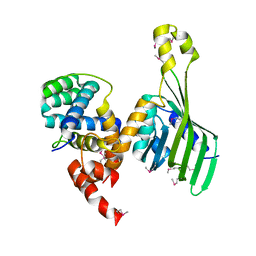 | | Structure of human LDB1 in complex with SSBP2 | | Descriptor: | LIM domain-binding protein 1, Single-stranded DNA-binding protein 2 | | Authors: | Wang, H, Wang, Z, Xu, W. | | Deposit date: | 2019-08-08 | | Release date: | 2020-01-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Crystal structure of human LDB1 in complex with SSBP2.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5IZ5
 
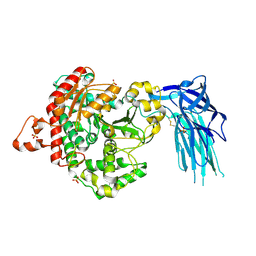 | | Human GIVD cytosolic phospholipase A2 | | Descriptor: | Cytosolic phospholipase A2 delta, SULFATE ION | | Authors: | Wang, H, Klein, M.G. | | Deposit date: | 2016-03-24 | | Release date: | 2016-06-08 | | Last modified: | 2016-06-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Human GIVD Cytosolic Phospholipase A2 Reveals Insights into Substrate Recognition.
J.Mol.Biol., 428, 2016
|
|
5IZR
 
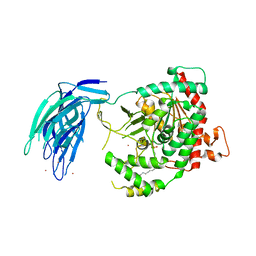 | | Human GIVD cytosolic phospholipase A2 in complex with Methyl gamma-Linolenyl Fluorophosphonate inhibitor and Terbium Chloride | | Descriptor: | Cytosolic phospholipase A2 delta, TERBIUM(III) ION, methyl (R)-(6Z,9Z,12Z)-octadeca-6,9,12-trien-1-ylphosphonofluoridate | | Authors: | Wang, H, Klein, M.G. | | Deposit date: | 2016-03-25 | | Release date: | 2016-06-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure of Human GIVD Cytosolic Phospholipase A2 Reveals Insights into Substrate Recognition.
J.Mol.Biol., 428, 2016
|
|
5J0F
 
 | | Monomeric Human Cu,Zn Superoxide dismutase, loops IV and VII deleted, apo form, circular permutant P4/5 | | Descriptor: | GLYCEROL, Superoxide dismutase [Cu-Zn],OXIDOREDUCTASE,Superoxide dismutase [Cu-Zn] | | Authors: | Wang, H, Lang, L, Logan, D, Danielsson, J, Oliveberg, M. | | Deposit date: | 2016-03-28 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Tricking a Protein To Swap Strands.
J. Am. Chem. Soc., 138, 2016
|
|
4MM4
 
 | |
4MM8
 
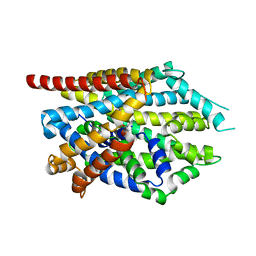 | |
4MMF
 
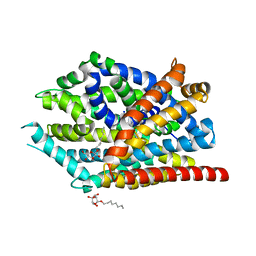 | | Crystal structure of LeuBAT (delta5 mutant) in complex with mazindol | | Descriptor: | (5R)-5-(4-chlorophenyl)-2,5-dihydro-3H-imidazo[2,1-a]isoindol-5-ol, SODIUM ION, Transporter, ... | | Authors: | Wang, H, Gouaux, E. | | Deposit date: | 2013-09-08 | | Release date: | 2013-10-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for action by diverse antidepressants on biogenic amine transporters.
Nature, 503, 2013
|
|
4MM9
 
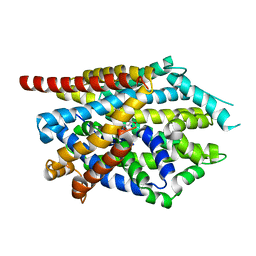 | |
4MM5
 
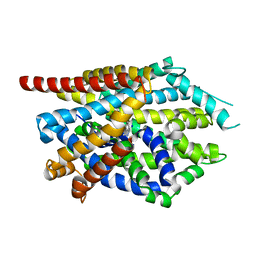 | | Crystal structure of LeuBAT (delta13 mutant) in complex with sertraline | | Descriptor: | (1S,4S)-4-(3,4-dichlorophenyl)-N-methyl-1,2,3,4-tetrahydronaphthalen-1-amine, SODIUM ION, Transporter | | Authors: | Wang, H, Gouaux, E. | | Deposit date: | 2013-09-08 | | Release date: | 2013-10-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for action by diverse antidepressants on biogenic amine transporters.
Nature, 503, 2013
|
|
4MMA
 
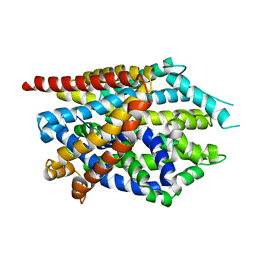 | |
4MMB
 
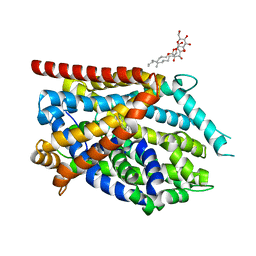 | | Crystal structure of LeuBAT (delta6 mutant) in complex with sertraline | | Descriptor: | (1S,4S)-4-(3,4-dichlorophenyl)-N-methyl-1,2,3,4-tetrahydronaphthalen-1-amine, SODIUM ION, Transporter, ... | | Authors: | Wang, H, Gouaux, E. | | Deposit date: | 2013-09-08 | | Release date: | 2013-10-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for action by diverse antidepressants on biogenic amine transporters.
Nature, 503, 2013
|
|
4MM6
 
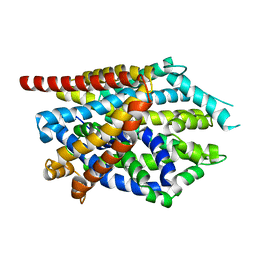 | |
4MME
 
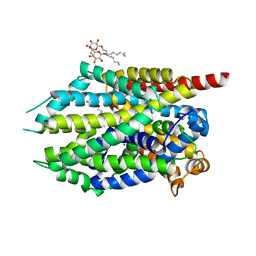 | | Crystal structure of LeuBAT (delta6 mutant) in complex with mazindol | | Descriptor: | (5R)-5-(4-chlorophenyl)-2,5-dihydro-3H-imidazo[2,1-a]isoindol-5-ol, SODIUM ION, Transporter, ... | | Authors: | Wang, H, Gouaux, E. | | Deposit date: | 2013-09-08 | | Release date: | 2013-10-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for action by diverse antidepressants on biogenic amine transporters.
Nature, 503, 2013
|
|
4MM7
 
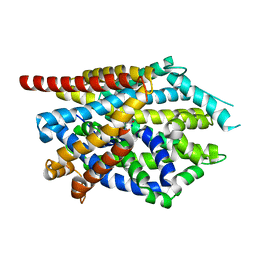 | |
4MMC
 
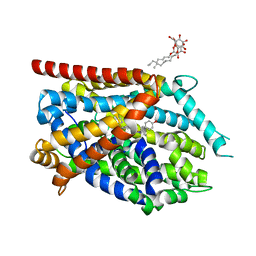 | |
4MMD
 
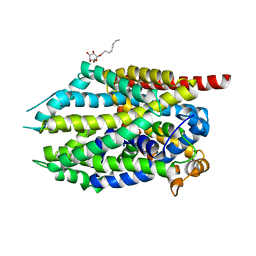 | |
6T9K
 
 | | SAGA Core module | | Descriptor: | Protein SPT3, SAGA-associated factor 73, Transcription factor SPT20, ... | | Authors: | Wang, H, Cheung, A, Cramer, P. | | Deposit date: | 2019-10-28 | | Release date: | 2020-01-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the transcription coactivator SAGA.
Nature, 577, 2020
|
|
6T9J
 
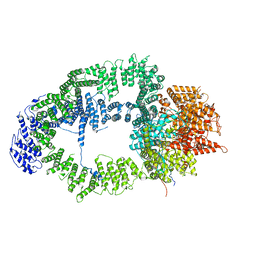 | | SAGA Tra1 module | | Descriptor: | Transcription factor SPT20, Transcription initiation factor TFIID subunit 12, Transcription-associated protein 1 | | Authors: | Wang, H, Cheung, A, Cramer, P. | | Deposit date: | 2019-10-28 | | Release date: | 2020-01-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the transcription coactivator SAGA.
Nature, 577, 2020
|
|
6T9I
 
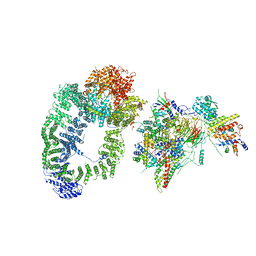 | | cryo-EM structure of transcription coactivator SAGA | | Descriptor: | Protein SPT3, SAGA-associated factor 73, Transcription factor SPT20, ... | | Authors: | Wang, H, Cheung, A, Cramer, P. | | Deposit date: | 2019-10-28 | | Release date: | 2020-01-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the transcription coactivator SAGA.
Nature, 577, 2020
|
|
6T9L
 
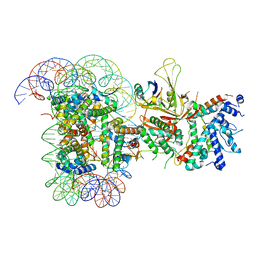 | |
3KH8
 
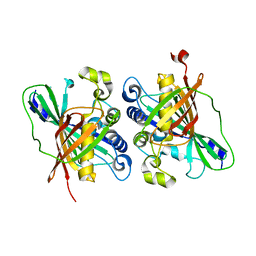 | | Crystal structure of MaoC-like dehydratase from Phytophthora Capsici | | Descriptor: | MaoC-like dehydratase | | Authors: | Wang, H, Zhang, K, Guo, J, Zhou, Q, Zheng, X, Sun, F, Pang, H, Zhang, X. | | Deposit date: | 2009-10-30 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of MaoC-like dehydratase from Phytophthora Capsici
To be Published
|
|
8EAV
 
 | | YAR027W and YAR028W in complex with c subunits from yeast VO complex | | Descriptor: | YAR027W or YAR028W, subunit from the c ring of yeast VO complex | | Authors: | Wang, H, Bueler, S.A, Rubinstein, J.L. | | Deposit date: | 2022-08-29 | | Release date: | 2022-11-02 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Structural basis of V-ATPase V O region assembly by Vma12p, 21p, and 22p.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EAT
 
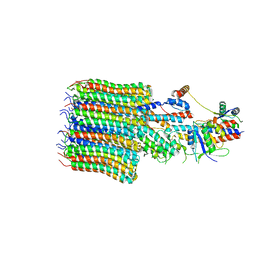 | | Yeast VO missing subunits a, e, and f in complex with Vma12-22p | | Descriptor: | V-type proton ATPase subunit F, V-type proton ATPase subunit c, V-type proton ATPase subunit c', ... | | Authors: | Wang, H, Bueler, S.A, Rubinstein, J.L. | | Deposit date: | 2022-08-29 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of V-ATPase V O region assembly by Vma12p, 21p, and 22p.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EAS
 
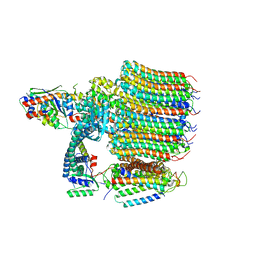 | | Yeast VO in complex with Vma12-22p | | Descriptor: | V-type proton ATPase assembly factor Vma12p, V-type proton ATPase subunit F, V-type proton ATPase subunit a, ... | | Authors: | Wang, H, Bueler, S.A, Rubinstein, J.L. | | Deposit date: | 2022-08-29 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis of V-ATPase V O region assembly by Vma12p, 21p, and 22p.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EAU
 
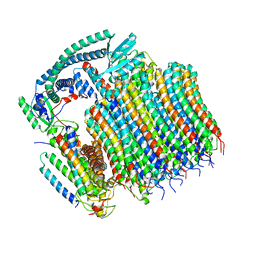 | | Yeast VO in complex with Vma21p | | Descriptor: | V-type proton ATPase subunit a, vacuolar isoform, V-type proton ATPase subunit c, ... | | Authors: | Wang, H, Bueler, S.A, Rubinstein, J.L. | | Deposit date: | 2022-08-29 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of V-ATPase V O region assembly by Vma12p, 21p, and 22p.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
