2ZY5
 
 | | R487A mutant of L-aspartate beta-decarboxylase | | Descriptor: | L-aspartate beta-decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Chen, H.-J, Ko, T.-P, Lee, C.-Y, Wang, N.-C, Wang, A.H.-J. | | Deposit date: | 2009-01-13 | | Release date: | 2009-01-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure, Assembly, and Mechanism of a PLP-Dependent Dodecameric l-Aspartate beta-Decarboxylase
Structure, 17, 2009
|
|
2ZY2
 
 | | dodecameric L-aspartate beta-decarboxylase | | Descriptor: | L-aspartate 4-carboxylyase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Chen, H.-J, Ko, T.-P, Lee, C.-Y, Wang, N.-C, Wang, A.H.-J. | | Deposit date: | 2009-01-13 | | Release date: | 2009-01-27 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure, Assembly, and Mechanism of a PLP-Dependent Dodecameric l-Aspartate beta-Decarboxylase
Structure, 17, 2009
|
|
2ZY3
 
 | | dodecameric L-aspartate beta-decarboxylase | | Descriptor: | L-aspartate beta-decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Chen, H.-J, Ko, T.-P, Lee, C.-Y, Wang, N.-C, Wang, A.H.-J. | | Deposit date: | 2009-01-13 | | Release date: | 2009-01-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure, Assembly, and Mechanism of a PLP-Dependent Dodecameric l-Aspartate beta-Decarboxylase
Structure, 17, 2009
|
|
1GVP
 
 | | GENE V PROTEIN (SINGLE-STRANDED DNA BINDING PROTEIN) | | Descriptor: | GENE V PROTEIN | | Authors: | Su, S, Gao, Y.-G, Zhang, H, Terwilliger, T.C, Wang, A.H.-J. | | Deposit date: | 1997-02-26 | | Release date: | 1997-09-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Analyses of the stability and function of three surface mutants (R82C, K69H, and L32R) of the gene V protein from Ff phage by X-ray crystallography.
Protein Sci., 6, 1997
|
|
1GKH
 
 | | MUTANT K69H OF GENE V PROTEIN (SINGLE-STRANDED DNA BINDING PROTEIN) | | Descriptor: | GENE V PROTEIN | | Authors: | Su, S, Gao, Y.-G, Zhang, H, Terwilliger, T.C, Wang, A.H.-J. | | Deposit date: | 1997-03-04 | | Release date: | 1997-09-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Analyses of the stability and function of three surface mutants (R82C, K69H, and L32R) of the gene V protein from Ff phage by X-ray crystallography.
Protein Sci., 6, 1997
|
|
1M7J
 
 | | Crystal structure of D-aminoacylase defines a novel subset of amidohydrolases | | Descriptor: | ACETATE ION, D-aminoacylase, ZINC ION | | Authors: | Liaw, S.-H, Chen, S.-J, Ko, T.-P, Hsu, C.-S, Wang, A.H.-J, Tsai, Y.-C. | | Deposit date: | 2002-07-22 | | Release date: | 2003-02-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of D-Aminoacylase from Alcaligenes faecalis DA1. A NOVEL SUBSET OF AMIDOHYDROLASES AND INSIGHTS INTO THE ENZYME MECHANISM.
J.Biol.Chem., 278, 2003
|
|
1I3W
 
 | | ACTINOMYCIN D BINDING TO CGATCGATCG | | Descriptor: | 5'-D(*C*GP*AP*TP*CP*GP*AP*(BRU)P*CP*GP)-3', ACTINOMYCIN D | | Authors: | Robinson, H, Gao, Y.-G, Yang, X.-L, Sanishvili, R, Joachimiak, A, Wang, A.H.-J. | | Deposit date: | 2001-02-17 | | Release date: | 2001-05-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallographic Analysis of a Novel Complex of Actinomycin D Bound to the DNA Decamer Cgatcgatcg.
Biochemistry, 40, 2001
|
|
2ZY4
 
 | | dodecameric L-aspartate beta-decarboxylase | | Descriptor: | CHLORIDE ION, L-aspartate beta-decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Chen, H.-J, Ko, T.-P, Lee, C.-Y, Wang, N.-C, Wang, A.H.-J. | | Deposit date: | 2009-01-13 | | Release date: | 2009-01-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure, Assembly, and Mechanism of a PLP-Dependent Dodecameric l-Aspartate beta-Decarboxylase
Structure, 17, 2009
|
|
1NDN
 
 | | MOLECULAR STRUCTURE OF NICKED DNA. MODEL T4 | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*AP*AP*CP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*GP*TP*T)-3'), DNA (5'-D(*TP*TP*CP*GP*CP*G)-3') | | Authors: | Aymani, J, Coll, M, Van Der Marel, G.A, Van Boom, J.H, Wang, A.H.-J, Rich, A. | | Deposit date: | 1992-01-15 | | Release date: | 1992-07-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular structure of nicked DNA: a substrate for DNA repair enzymes.
Proc.Natl.Acad.Sci.USA, 87, 1990
|
|
1FD5
 
 | | BINDING OF A MACROCYCLIC BISACRIDINE AND AMETANTRONE TO CGTACG INVOLVES SIMILAR UNUSUAL INTERCALATION PLATFORMS (BISACRIDINE COMPLEX) | | Descriptor: | COBALT (II) ION, DNA (5'-D(*CP*GP*TP*AP*CP*G)-3'), MACROCYCLIC-BIS-9-AMINO-ACRIDINE, ... | | Authors: | Yang, X.-L, Robinson, H, Gao, Y.-G, Wang, A.H.-J. | | Deposit date: | 2000-07-19 | | Release date: | 2000-08-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Binding of a macrocyclic bisacridine and ametantrone to CGTACG involves similar unusual intercalation platforms.
Biochemistry, 39, 2000
|
|
1FDG
 
 | | BINDING OF A MACROCYCLIC BISACRIDINE AND AMETANTRONE TO CGTACG INVOLVES SIMILAR UNUSUAL INTERCALATION PLATFORMS (AMETANTRONE COMPLEX) | | Descriptor: | 1,4-BIS-[2-(2-HYDROXY-ETHYLAMINO)-ETHYLAMINO]-ANTHRAQUINONE, COBALT (II) ION, DNA (5'-D(*CP*GP*TP*AP*CP*G)-3'), ... | | Authors: | Yang, X.-L, Robinson, H, Gao, Y.-G, Wang, A.H.-J. | | Deposit date: | 2000-07-20 | | Release date: | 2000-08-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Binding of a macrocyclic bisacridine and ametantrone to CGTACG involves similar unusual intercalation platforms.
Biochemistry, 39, 2000
|
|
1FY2
 
 | | Aspartyl Dipeptidase | | Descriptor: | ASPARTYL DIPEPTIDASE, CADMIUM ION | | Authors: | Hakansson, K, Wang, A.H.-J, Miller, C.G. | | Deposit date: | 2000-09-28 | | Release date: | 2001-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The structure of aspartyl dipeptidase reveals a unique fold with a Ser-His-Glu catalytic triad.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1FYE
 
 | |
1YHB
 
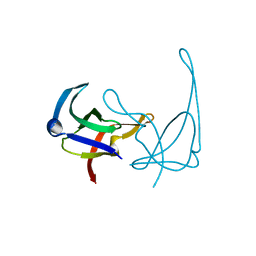 | | CRYSTAL STRUCTURES OF Y41H AND Y41F MUTANTS OF GENE V PROTEIN FROM FF PHAGE SUGGEST POSSIBLE PROTEIN-PROTEIN INTERACTIONS IN GVP-SSDNA COMPLEX | | Descriptor: | GENE V PROTEIN | | Authors: | Guan, Y, Zhang, H, Konings, R.N.H, Hilbers, C.W, Terwilliger, T.C, Wang, A.H.-J. | | Deposit date: | 1994-04-14 | | Release date: | 1994-06-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of Y41H and Y41F mutants of gene V protein from Ff phage suggest possible protein-protein interactions in the GVP-ssDNA complex.
Biochemistry, 33, 1994
|
|
8KB6
 
 | | Crystal Structure of Canine TNF-alpha | | Descriptor: | Tumor necrosis factor | | Authors: | Lee, C.C, Wang, A.H.-J. | | Deposit date: | 2023-08-03 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.850166 Å) | | Cite: | Structure-based development of a canine TNF-alpha-specific antibody using adalimumab as a template.
Protein Sci., 33, 2024
|
|
1YHA
 
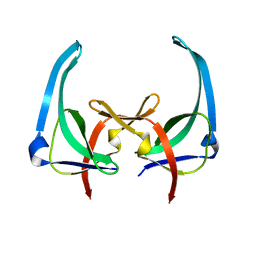 | | CRYSTAL STRUCTURES OF Y41H AND Y41F MUTANTS OF GENE V PROTEIN FROM FF PHAGE SUGGEST POSSIBLE PROTEIN-PROTEIN INTERACTIONS IN GVP-SSDNA COMPLEX | | Descriptor: | GENE V PROTEIN | | Authors: | Guan, Y, Zhang, H, Konings, R.N.H, Hilbers, C.W, Terwilliger, T.C, Wang, A.H.-J. | | Deposit date: | 1994-04-14 | | Release date: | 1994-06-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of Y41H and Y41F mutants of gene V protein from Ff phage suggest possible protein-protein interactions in the GVP-ssDNA complex.
Biochemistry, 33, 1994
|
|
1VTE
 
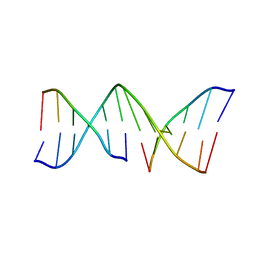 | | MOLECULAR STRUCTURE OF NICKED DNA. MODEL A4 | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*AP*AP*CP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*GP*TP*T)-3'), DNA (5'-D(*TP*TP*CP*GP*CP*G)-3') | | Authors: | Aymani, J, Coll, M, Van Der Marel, G.A, Van Boom, J.H, Wang, A.H.-J, Rich, A. | | Deposit date: | 1990-05-21 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular structure of nicked DNA: a substrate for DNA repair enzymes.
Proc. Natl. Acad. Sci. U.S.A., 87, 1990
|
|
1WL2
 
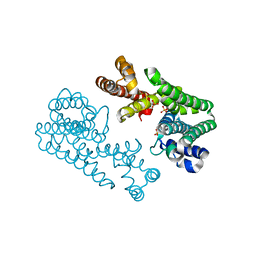 | | Crystal Structure Of Octaprenyl Pyrophosphate Synthase From Hyperthermophilic Thermotoga Maritima R90A mutant | | Descriptor: | SULFATE ION, octoprenyl-diphosphate synthase | | Authors: | Guo, R.T, Kuo, C.J, Cheng, Y.S, Cheng, Y.L, Liang, P.H, Wang, A.H.-J. | | Deposit date: | 2004-06-18 | | Release date: | 2005-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Biochemical and Structural Basis for Octaprenyl Pyrophosphate Synthase
To be Published
|
|
1WL1
 
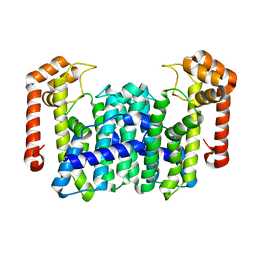 | | Crystal Structure Of Octaprenyl Pyrophosphate Synthase From Hyperthermophilic Thermotoga Maritima H74A mutant | | Descriptor: | octoprenyl-diphosphate synthase | | Authors: | Guo, R.T, Kuo, C.J, Cheng, Y.S, Cheng, Y.L, Liang, P.H, Wang, A.H.-J. | | Deposit date: | 2004-06-18 | | Release date: | 2005-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Biochemical and Structural Basis for Octaprenyl Pyrophosphate Synthase
To be Published
|
|
1WL0
 
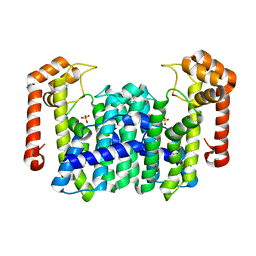 | | Crystal Structure Of Octaprenyl Pyrophosphate Synthase From Hyperthermophilic Thermotoga Maritima R44A mutant | | Descriptor: | SULFATE ION, octoprenyl-diphosphate synthase | | Authors: | Guo, R.T, Kuo, C.J, Cheng, Y.S, Cheng, Y.L, Liang, P.H, Wang, A.H.-J. | | Deposit date: | 2004-06-18 | | Release date: | 2005-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Biochemical and Structural Basis for Octaprenyl Pyrophosphate Synthase
To be Published
|
|
1MNV
 
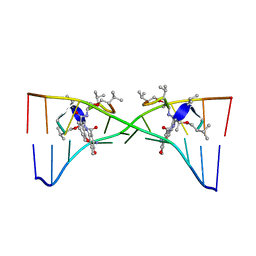 | | Actinomycin D binding to ATGCTGCAT | | Descriptor: | 5'-D(*AP*TP*GP*CP*TP*GP*CP*AP*T)-3', ACTINOMYCIN D | | Authors: | Hou, M.-H, Robinson, H, Gao, Y.-G, Wang, A.H.-J. | | Deposit date: | 2002-09-06 | | Release date: | 2002-11-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Actinomycin D Bound to the Ctg Triplet Repeat Sequences Linked to Neurological Diseases
Nucleic Acids Res., 30, 2002
|
|
3MBR
 
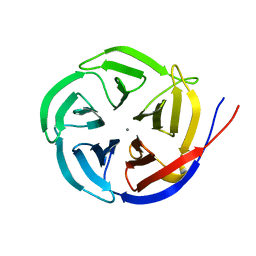 | | Crystal Structure of the Glutaminyl Cyclase from Xanthomonas campestris | | Descriptor: | CALCIUM ION, Glutamine cyclotransferase | | Authors: | Huang, W.-L, Wang, Y.-R, Ko, T.-P, Chia, C.-Y, Huang, K.-F, Wang, A.H.-J. | | Deposit date: | 2010-03-25 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Crystal structure and functional analysis of the glutaminyl cyclase from Xanthomonas campestris
J.Mol.Biol., 401, 2010
|
|
2ZXA
 
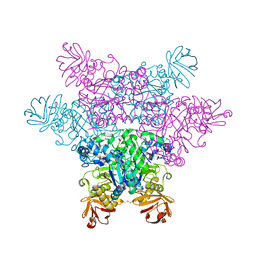 | | alpha-L-fucosidase complexed with inhibitor, FNJ-acetyl | | Descriptor: | Alpha-L-fucosidase, putative, N-{[(2R,3R,4R,5R,6S)-3,4,5-trihydroxy-6-methylpiperidin-2-yl]methyl}acetamide | | Authors: | Wu, H.-J, Ko, T.-P, Ho, C.-W, Lin, C.-H, Wang, A.H.-J. | | Deposit date: | 2008-12-22 | | Release date: | 2009-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural basis of alpha-fucosidase inhibition by iminocyclitols with K(i) values in the micro- to picomolar range.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
2ZX5
 
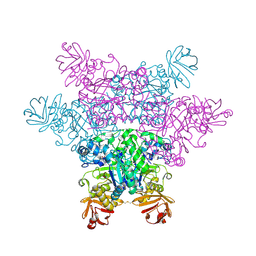 | | alpha-L-fucosidase complexed with inhibitor, F10 | | Descriptor: | 3-(1H-indol-3-yl)-N-{[(2R,3R,4R,5R,6S)-3,4,5-trihydroxy-6-methylpiperidin-2-yl]methyl}propanamide, Alpha-L-fucosidase, putative | | Authors: | Wu, H.-J, Ko, T.-P, Ho, C.-W, Lin, C.-H, Wang, A.H.-J. | | Deposit date: | 2008-12-19 | | Release date: | 2009-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis of alpha-fucosidase inhibition by iminocyclitols with K(i) values in the micro- to picomolar range.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
1UCS
 
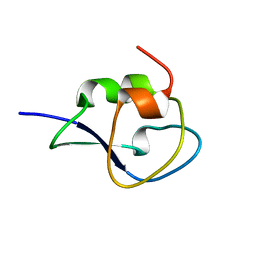 | | Type III Antifreeze Protein RD1 from an Antarctic Eel Pout | | Descriptor: | Antifreeze peptide RD1 | | Authors: | Ko, T.-P, Robinson, H, Gao, Y.-G, Cheng, C.-H.C, DeVries, A.L, Wang, A.H.-J. | | Deposit date: | 2003-04-21 | | Release date: | 2003-05-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (0.62 Å) | | Cite: | The refined crystal structure of an eel pout type III antifreeze protein RD1 at 0.62-A resolution reveals structural microheterogeneity of protein and solvation.
Biophys.J., 84, 2003
|
|
