8IQL
 
 | | Structural basis of the specificity and interaction mechanism of Bmf binding to pro-survival proteins | | Descriptor: | Apoptosis regulator Bcl-2, Bcl-2-modifying factor | | Authors: | Wang, H, Guo, M, Wei, H, Chen, Y. | | Deposit date: | 2023-03-16 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9577 Å) | | Cite: | Structural basis of the specificity and interaction mechanism of Bmf binding to pro-survival Bcl-2 family proteins.
Comput Struct Biotechnol J, 21, 2023
|
|
6K8O
 
 | | Crystal structure of the Sulfolobus solfataricus topoisomerase III in complex with DNA | | Descriptor: | DNA (5'-D(*GP*CP*AP*AP*GP*GP*TP*C)-3'), ZINC ION, topoisomerase III | | Authors: | Wang, H.Q, Zhang, J.H, Zheng, X, Zheng, Z.F, Dong, Y.H, Huang, L, Gong, Y. | | Deposit date: | 2019-06-13 | | Release date: | 2020-06-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the Sulfolobus solfataricus topoisomerase III reveal that its C-terminal novel zinc finger part is a unique decatenation domain
To Be Published
|
|
2PW3
 
 | | Structure of the PDE4D-cAMP complex | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, ZINC ION, cAMP-specific 3',5'-cyclic phosphodiesterase 4D | | Authors: | Wang, H, Robinson, H, Ke, H. | | Deposit date: | 2007-05-10 | | Release date: | 2007-10-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | The molecular basis for different recognition of substrates by phosphodiesterase families 4 and 10.
J.Mol.Biol., 371, 2007
|
|
8HIB
 
 | |
6HBC
 
 | | Structure of the repeat unit in the network formed by CcmM and Rubisco from Synechococcus elongatus | | Descriptor: | Carbon dioxide concentrating mechanism protein CcmM, Ribulose 1,5-bisphosphate carboxylase small subunit, Ribulose bisphosphate carboxylase large chain | | Authors: | Wang, H, Yan, X, Aigner, H, Bracher, A, Nguyen, N.D, Hee, W.Y, Long, B.M, Price, G.D, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2018-08-10 | | Release date: | 2018-12-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Rubisco condensate formation by CcmM in beta-carboxysome biogenesis.
Nature, 566, 2019
|
|
5YXA
 
 | | Crystal structure of the C-terminal fragment of NS1 protein from yellow fever virus | | Descriptor: | Non-structural protein 1 | | Authors: | Wang, H, Song, H, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2017-12-04 | | Release date: | 2018-01-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the C-terminal fragment of NS1 protein from yellow fever virus.
Sci China Life Sci, 60, 2017
|
|
8G9E
 
 | |
6I6O
 
 | | Circular permutant of ribosomal protein S6, swap helix 2, L75A mutant | | Descriptor: | 30S ribosomal protein S6,30S ribosomal protein S6 | | Authors: | Wang, H, Logan, D.T, Oliveberg, M. | | Deposit date: | 2018-11-15 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Exposing the distinctive modular behavior of beta-strands and alpha-helices in folded proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7U2K
 
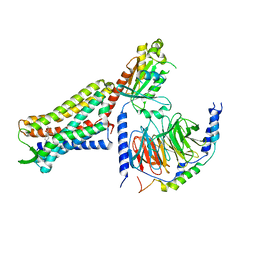 | | C6-guano bound Mu Opioid Receptor-Gi Protein Complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Wang, H, Kobilka, B. | | Deposit date: | 2022-02-24 | | Release date: | 2022-12-07 | | Last modified: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure-based design of bitopic ligands for the μ-opioid receptor.
Nature, 613, 2023
|
|
6I6U
 
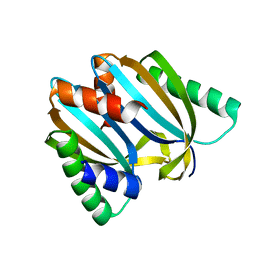 | | Circular permutant of ribosomal protein S6, adding 9aa to N terminal of P81-82, L75A mutant | | Descriptor: | 30S ribosomal protein S6,30S ribosomal protein S6 | | Authors: | Wang, H, Logan, D.T, Oliveberg, M. | | Deposit date: | 2018-11-15 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Exposing the distinctive modular behavior of beta-strands and alpha-helices in folded proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6I6W
 
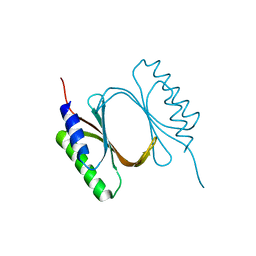 | |
6ISB
 
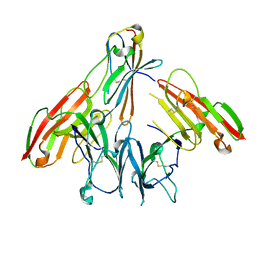 | | crystal structure of human CD226 | | Descriptor: | CD226 antigen | | Authors: | Wang, H, Qi, J, Zhang, S, Li, Y, Tan, S, Gao, G.F. | | Deposit date: | 2018-11-16 | | Release date: | 2018-12-26 | | Last modified: | 2019-02-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Binding mode of the side-by-side two-IgV molecule CD226/DNAM-1 to its ligand CD155/Necl-5.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6ISA
 
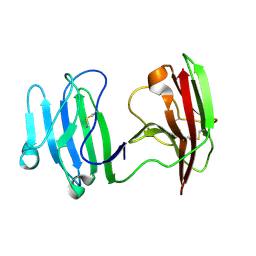 | | mCD226 | | Descriptor: | CD226 antigen | | Authors: | Wang, H, Qi, J, Zhang, S, Li, Y, Tan, S, Gao, G.F. | | Deposit date: | 2018-11-16 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding mode of the side-by-side two-IgV molecule CD226/DNAM-1 to its ligand CD155/Necl-5.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6I6S
 
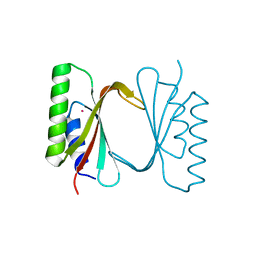 | | Circular permutant of ribosomal protein S6, adding 9aa to C terminal of P68-69, L75A mutant | | Descriptor: | 30S ribosomal protein S6,30S ribosomal protein S6,30S ribosomal protein S6,30S ribosomal protein S6,30S ribosomal protein S6,30S ribosomal protein S6,30S ribosomal protein S6,30S ribosomal protein S6, POTASSIUM ION, SODIUM ION | | Authors: | Wang, H, Logan, D.T, Oliveberg, M. | | Deposit date: | 2018-11-15 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Exposing the distinctive modular behavior of beta-strands and alpha-helices in folded proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6ISC
 
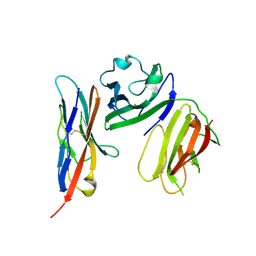 | | complex structure of mCD226-ecto and hCD155-D1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CD226 antigen, Poliovirus receptor | | Authors: | Wang, H, Qi, J, Zhang, S, Li, Y, Tan, S, Gao, G.F. | | Deposit date: | 2018-11-16 | | Release date: | 2018-12-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Binding mode of the side-by-side two-IgV molecule CD226/DNAM-1 to its ligand CD155/Necl-5.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6I6E
 
 | |
6I69
 
 | |
6I6I
 
 | | Circular permutant of ribosomal protein S6, adding 6aa to C terminal of P68-69, L75A mutant | | Descriptor: | 30S ribosomal protein S6,30S ribosomal protein S6, SULFATE ION | | Authors: | Wang, H, Logan, D.T, Oliveberg, M. | | Deposit date: | 2018-11-15 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Exposing the distinctive modular behavior of beta-strands and alpha-helices in folded proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6I6Y
 
 | |
7BFG
 
 | |
7BFC
 
 | |
7BFD
 
 | |
7BFF
 
 | |
7BFE
 
 | |
1WWI
 
 | | Crystal structure of ttk003001566 from Thermus Thermophilus HB8 | | Descriptor: | hypothetical protein TTHA1479 | | Authors: | Wang, H, Murayama, K, Terada, T, Chen, L, Liu, Z.J, Wang, B.C, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-01-05 | | Release date: | 2005-07-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of ttk003001566 from Thermus Thermophilus HB8
TO BE PUBLISHED
|
|
