2CSL
 
 | | Crystal structure of TTHA0137 from Thermus Thermophilus HB8 | | Descriptor: | protein translation initiation inhibitor | | Authors: | Wang, H, Murayama, K, Terada, T, Chen, L, Jin, Z, Chrzas, J, Liu, Z.J, Wang, B.C, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-22 | | Release date: | 2005-11-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of TTHA0137 from Thermus Thermophilus HB8
To be Published
|
|
3VK0
 
 | | Crystal Structure of hypothetical transcription factor NHTF from Neisseria | | Descriptor: | Transcriptional regulator | | Authors: | Wang, H.-C, Ko, T.-P, Wu, M.-L, Wu, H.-J, Ku, S.-C, Wang, A.H.-J. | | Deposit date: | 2011-11-01 | | Release date: | 2012-03-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Neisseria conserved protein DMP19 is a DNA mimic protein that prevents DNA binding to a hypothetical nitrogen-response transcription factor
Nucleic Acids Res., 2012
|
|
3VJZ
 
 | | Crystal structure of the DNA mimic protein DMP19 | | Descriptor: | Putative uncharacterized protein | | Authors: | Wang, H.-C, Ko, T.-P, Wang, A.H.-J. | | Deposit date: | 2011-11-01 | | Release date: | 2012-03-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Neisseria conserved protein DMP19 is a DNA mimic protein that prevents DNA binding to a hypothetical nitrogen-response transcription factor
Nucleic Acids Res., 2012
|
|
3W1O
 
 | | Neisseria DNA mimic protein DMP12 | | Descriptor: | DNA mimic protein DMP12, MAGNESIUM ION | | Authors: | Wang, H.C, Ko, T.P, Wu, M.L, Wang, A.H.J. | | Deposit date: | 2012-11-19 | | Release date: | 2013-04-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Neisseria conserved hypothetical protein DMP12 is a DNA mimic that binds to histone-like HU protein
Nucleic Acids Res., 41, 2013
|
|
1BM5
 
 | |
8R0F
 
 | | Capsid structure of Giardiavirus (GLV) HP strain | | Descriptor: | Capsid protein | | Authors: | Wang, H, Gianluca, M, Munke, A, Hassan, M.M, Lalle, M, Okamoto, K. | | Deposit date: | 2023-10-31 | | Release date: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.14 Å) | | Cite: | Capsid structure of Giardiavirus (GLV) HP strain
To Be Published
|
|
8R0G
 
 | | Capsid structure of Giardiavirus (GLV) CAT strain | | Descriptor: | Capsid protein | | Authors: | Wang, H, Gianluca, M, Munke, A, Hassan, M.M, Lalle, M, Okamoto, K. | | Deposit date: | 2023-10-31 | | Release date: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Capsid structure of Giardiavirus (GLV) CAT strain
To Be Published
|
|
5C3I
 
 | | Crystal structure of the quaternary complex of histone H3-H4 heterodimer with chaperone ASF1 and the replicative helicase subunit MCM2 | | Descriptor: | DNA replication licensing factor MCM2,MCM2, Histone H3.1, Histone H4, ... | | Authors: | Wang, H, Wang, M, Yang, N, Xu, R.M. | | Deposit date: | 2015-06-17 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the quaternary complex of histone H3-H4 heterodimer with chaperone ASF1 and the replicative helicase subunit MCM2
Protein Cell, 6, 2015
|
|
7ZSB
 
 | |
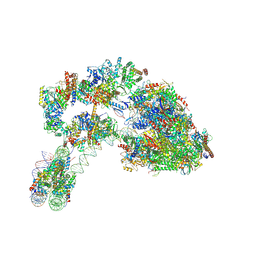
7ZSA
 
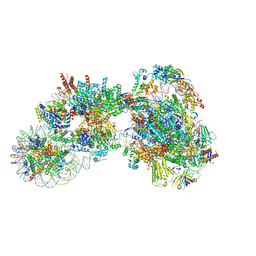 | |
7ZS9
 
 | |
7YRN
 
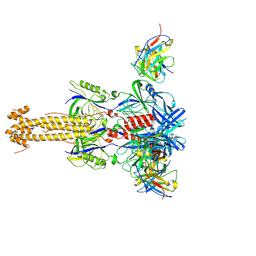 | |
3WDF
 
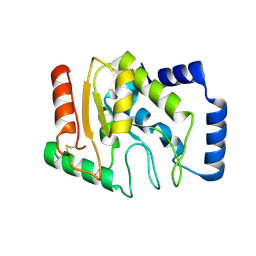 | | Staphylococcus aureus UDG | | Descriptor: | Uracil-DNA glycosylase | | Authors: | Wang, H.C, Ko, T.P, Wang, A.H.J. | | Deposit date: | 2013-06-18 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Staphylococcus aureus protein SAUGI acts as a uracil-DNA glycosylase inhibitor.
Nucleic Acids Res., 42, 2013
|
|
3WDG
 
 | | Staphylococcus aureus UDG / UGI complex | | Descriptor: | Uncharacterized protein, Uracil-DNA glycosylase | | Authors: | Wang, H.C, Ko, T.P, Wang, A.H.J. | | Deposit date: | 2013-06-18 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Staphylococcus aureus protein SAUGI acts as a uracil-DNA glycosylase inhibitor.
Nucleic Acids Res., 42, 2013
|
|
2DY1
 
 | | Crystal structure of EF-G-2 from Thermus thermophilus | | Descriptor: | Elongation factor G, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Wang, H, Takemoto, C, Murayama, K, Terada, T, Chen, L, Liu, Z.J, Wang, B.C, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-09-04 | | Release date: | 2007-09-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of EF-G-2 from Thermus thermophilus
To be Published
|
|
6LHE
 
 | | Crystal Structure of Gold-bound NDM-1 | | Descriptor: | GOLD ION, Metallo-beta-lactamase type 2, SULFATE ION | | Authors: | Wang, H, Sun, H, Wang, M. | | Deposit date: | 2019-12-07 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.206 Å) | | Cite: | Resensitizing carbapenem- and colistin-resistant bacteria to antibiotics using auranofin.
Nat Commun, 11, 2020
|
|
8EAV
 
 | | YAR027W and YAR028W in complex with c subunits from yeast VO complex | | Descriptor: | YAR027W or YAR028W, subunit from the c ring of yeast VO complex | | Authors: | Wang, H, Bueler, S.A, Rubinstein, J.L. | | Deposit date: | 2022-08-29 | | Release date: | 2022-11-02 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Structural basis of V-ATPase V O region assembly by Vma12p, 21p, and 22p.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EAT
 
 | | Yeast VO missing subunits a, e, and f in complex with Vma12-22p | | Descriptor: | V-type proton ATPase subunit F, V-type proton ATPase subunit c, V-type proton ATPase subunit c', ... | | Authors: | Wang, H, Bueler, S.A, Rubinstein, J.L. | | Deposit date: | 2022-08-29 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of V-ATPase V O region assembly by Vma12p, 21p, and 22p.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6Z1G
 
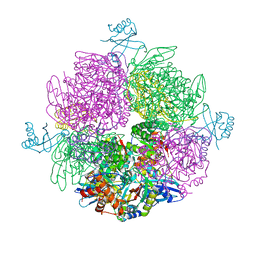 | | CryoEM structure of the interaction between Rubisco Activase small-subunit-like (SSUL) domain with Rubisco from Nostoc sp. (strain PCC7120) | | Descriptor: | Ribulose bisphosphate carboxylase large chain, Ribulose bisphosphate carboxylase small chain, Ribulose bisphosphate carboxylase/oxygenase activase | | Authors: | Wang, H, Bracher, A, Flecken, M, Popilka, L, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2020-05-13 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | Dual Functions of a Rubisco Activase in Metabolic Repair and Recruitment to Carboxysomes.
Cell, 183, 2020
|
|
8EAS
 
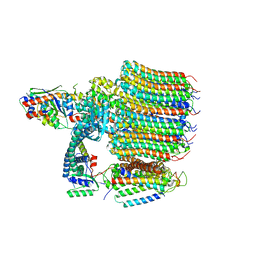 | | Yeast VO in complex with Vma12-22p | | Descriptor: | V-type proton ATPase assembly factor Vma12p, V-type proton ATPase subunit F, V-type proton ATPase subunit a, ... | | Authors: | Wang, H, Bueler, S.A, Rubinstein, J.L. | | Deposit date: | 2022-08-29 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis of V-ATPase V O region assembly by Vma12p, 21p, and 22p.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EAU
 
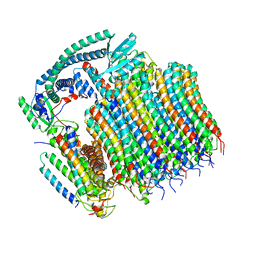 | | Yeast VO in complex with Vma21p | | Descriptor: | V-type proton ATPase subunit a, vacuolar isoform, V-type proton ATPase subunit c, ... | | Authors: | Wang, H, Bueler, S.A, Rubinstein, J.L. | | Deposit date: | 2022-08-29 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of V-ATPase V O region assembly by Vma12p, 21p, and 22p.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
1BPR
 
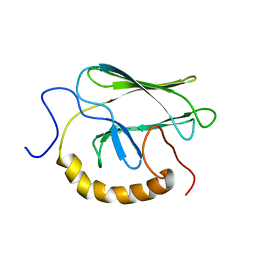 | | NMR STRUCTURE OF THE SUBSTRATE BINDING DOMAIN OF DNAK, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DNAK | | Authors: | Wang, H, Kurochkin, A.V, Pang, Y, Hu, W, Flynn, G.C, Zuiderweg, E.R.P. | | Deposit date: | 1998-08-11 | | Release date: | 1999-03-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the 21 kDa chaperone protein DnaK substrate binding domain: a preview of chaperone-protein interaction.
Biochemistry, 37, 1998
|
|
7B8V
 
 | |
1WWI
 
 | | Crystal structure of ttk003001566 from Thermus Thermophilus HB8 | | Descriptor: | hypothetical protein TTHA1479 | | Authors: | Wang, H, Murayama, K, Terada, T, Chen, L, Liu, Z.J, Wang, B.C, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-01-05 | | Release date: | 2005-07-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of ttk003001566 from Thermus Thermophilus HB8
TO BE PUBLISHED
|
|
6O60
 
 | | Crystal structure of GGTase3-FBXL2-SKP1 complex | | Descriptor: | F-box/LRR-repeat protein 2, Geranylgeranyl transferase type-2 subunit beta, Protein prenyltransferase alpha subunit repeat-containing protein 1, ... | | Authors: | Wang, H, Zheng, N. | | Deposit date: | 2019-03-04 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | GGTase3 is a newly identified geranylgeranyltransferase targeting a ubiquitin ligase.
Nat.Struct.Mol.Biol., 26, 2019
|
|
