8XPV
 
 | | The Crystal Structure of EphA2 from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, 1-(3,3-dimethylbutyl)-3-{2-fluoro-4-methyl-5-[7-methyl-2-(methylamino)pyrido[2,3-d]pyrimidin-6-yl]phenyl}urea, Ephrin type-A receptor 2, ... | | Authors: | Wang, F, Cheng, W, Lv, Z, Meng, Q, Xu, Y. | | Deposit date: | 2024-01-04 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Crystal Structure of EphA2 from Biortus.
To Be Published
|
|
4JJN
 
 | | Crystal structure of heterochromatin protein Sir3 in complex with a silenced yeast nucleosome | | Descriptor: | DNA (146-MER), Histone H2A.2, Histone H2B.2, ... | | Authors: | Wang, F, Li, G, Mohammed, A, Lu, C, Currie, M, Johnson, A, Moazed, D. | | Deposit date: | 2013-03-08 | | Release date: | 2013-05-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Heterochromatin protein Sir3 induces contacts between the amino terminus of histone H4 and nucleosomal DNA.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
8XFY
 
 | | The Crystal Structure of RSK2 from Biortus. | | Descriptor: | 2,6-bis(fluoranyl)-4-[4-(4-morpholin-4-ylphenyl)pyridin-3-yl]phenol, Ribosomal protein S6 kinase alpha-3 | | Authors: | Wang, F, Cheng, W, Lv, Z, Lin, D, Pan, W. | | Deposit date: | 2023-12-14 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Crystal Structure of RSK2 from Biortus.
To Be Published
|
|
7E9V
 
 | | The Crystal Structure of human UMP-CMP kinase from Biortus. | | Descriptor: | SULFATE ION, UMP-CMP kinase | | Authors: | Wang, F, Lin, D, Wang, R, Wei, X, Shen, Z, Wang, M. | | Deposit date: | 2021-03-05 | | Release date: | 2021-03-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of human UMP-CMP kinase from Biortus.
To Be Published
|
|
4L2Z
 
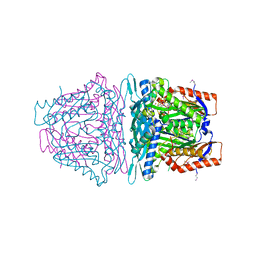 | | Crystal structure of S-Adenosylmethionine synthetase from Sulfolobus solfataricus complexed with SAE and PPi | | Descriptor: | DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Wang, F, Hurley, K.A, Helmich, K.E, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-06-05 | | Release date: | 2013-06-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.494 Å) | | Cite: | Understanding molecular recognition of promiscuity of thermophilic methionine adenosyltransferase sMAT from Sulfolobus solfataricus.
Febs J., 281, 2014
|
|
7VZT
 
 | | A human neutralizing antibody targeting SARS-CoV-2 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GH12-Heavy, GH12-LIGHT, ... | | Authors: | Wang, F.Z, Wang, Y, Tan, X.W, Shi, R, Yan, J.H. | | Deposit date: | 2021-11-16 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | GH12, glycosylation function
To Be Published
|
|
4M83
 
 | | Ensemble refinement of protein crystal structure (2IYF) of macrolide glycosyltransferases OleD complexed with UDP and Erythromycin A | | Descriptor: | ERYTHROMYCIN A, MAGNESIUM ION, Oleandomycin glycosyltransferase, ... | | Authors: | Wang, F, Helmich, K.E, Xu, W, Singh, S, Olmos Jr, J.L, Martinez iii, E, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-08-12 | | Release date: | 2013-09-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Crystal structure of macrolide glycosyltransferases OleD
To be Published
|
|
4M7P
 
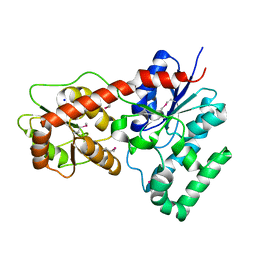 | | Ensemble refinement of protein crystal structure of macrolide glycosyltransferases OleD | | Descriptor: | Oleandomycin glycosyltransferase, SODIUM ION | | Authors: | Wang, F, Helmich, K.E, Xu, W, Singh, S, Olmos Jr, J.L, Martinez iii, E, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-08-12 | | Release date: | 2013-09-11 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of macrolide glycosyltransferases OleD
To be Published
|
|
4J1U
 
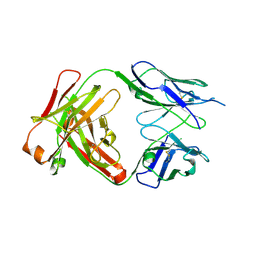 | | Crystal structure of antibody 93F3 unstable variant | | Descriptor: | antibody 93F3 Heavy chain, antibody 93F3 Light chain | | Authors: | Wang, F. | | Deposit date: | 2013-02-02 | | Release date: | 2013-03-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Somatic hypermutation maintains antibody thermodynamic stability during affinity maturation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7KHD
 
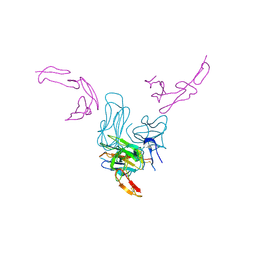 | | Human GITR-GITRL complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Tumor necrosis factor ligand superfamily member 18, Tumor necrosis factor receptor superfamily member 18 | | Authors: | Wang, F, Chau, B, West, S.M, Strop, P. | | Deposit date: | 2020-10-21 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.956102 Å) | | Cite: | Structures of mouse and human GITR-GITRL complexes reveal unique TNF superfamily interactions.
Nat Commun, 12, 2021
|
|
7KHX
 
 | | Mouse GITR-GITRL complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Tumor necrosis factor ligand superfamily member 18, ... | | Authors: | Wang, F, Chau, B, West, S.M, Strop, P. | | Deposit date: | 2020-10-22 | | Release date: | 2021-03-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.20569849 Å) | | Cite: | Structures of mouse and human GITR-GITRL complexes reveal unique TNF superfamily interactions.
Nat Commun, 12, 2021
|
|
8WF7
 
 | | The Crystal Structure of integrase from Biortus | | Descriptor: | ACETATE ION, Integrase, SULFATE ION | | Authors: | Wang, F, Cheng, W, Yuan, Z, Qi, J, Li, J. | | Deposit date: | 2023-09-19 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Crystal Structure of integrase from Biortus
To Be Published
|
|
8WD2
 
 | | The Crystal Structure of p53 from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, Cellular tumor antigen p53, PHOSPHATE ION, ... | | Authors: | Wang, F, Cheng, W, Yuan, Z, Qi, J, Lu, Y. | | Deposit date: | 2023-09-14 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Crystal Structure of p53 from Biortus.
To Be Published
|
|
8WF4
 
 | | The Crystal Structure of RSK1 from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, Ribosomal protein S6 kinase alpha-1 | | Authors: | Wang, F, Cheng, W, Lv, Z, Qi, J, Li, J. | | Deposit date: | 2023-09-19 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The Crystal Structure of RSK1 from Biortus.
To Be Published
|
|
8WFY
 
 | | The Crystal Structure of SHP2 from Biortus. | | Descriptor: | 6-(4-azanyl-4-methyl-piperidin-1-yl)-3-[2,3-bis(chloranyl)phenyl]pyrazin-2-amine, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Wang, F, Cheng, W, Yuan, Z, Qi, J, Li, J. | | Deposit date: | 2023-09-20 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Crystal Structure of SHP2 from Biortus.
To Be Published
|
|
8WD3
 
 | | The Crystal Structure of JMJD2A(M1-L359) from Biortus. | | Descriptor: | Lysine-specific demethylase 4A, NICKEL (II) ION, ZINC ION | | Authors: | Wang, F, Cheng, W, Lv, Z, Ju, C, Bao, C. | | Deposit date: | 2023-09-14 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The Crystal Structure of JMJD2A(M1-L359) from Biortus.
To Be Published
|
|
8WFE
 
 | | The Crystal Structure of PPARg from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Peroxisome proliferator-activated receptor gamma | | Authors: | Wang, F, Cheng, W, Lv, Z, Guo, S, Lin, D. | | Deposit date: | 2023-09-19 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Crystal Structure of PPARg from Biortus.
To Be Published
|
|
8WFR
 
 | | The Crystal Structure of PCSK9 from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Wang, F, Cheng, W, Lv, Z, Meng, Q, Lu, Y. | | Deposit date: | 2023-09-20 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Crystal Structure of PCSK9 from Biortus.
To Be Published
|
|
8XOY
 
 | | The Crystal Structure of PTP1B from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Wang, F, Cheng, W, Lv, Z, Meng, Q, Li, J. | | Deposit date: | 2024-01-02 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Crystal Structure of PTP1B from Biortus.
To Be Published
|
|
8WSW
 
 | | The Crystal Structure of LIMK2a from Biortus | | Descriptor: | 1,2-ETHANEDIOL, LIM domain kinase 2, ~{N}-[5-[2-[2,6-bis(chloranyl)phenyl]-5-[bis(fluoranyl)methyl]pyrazol-3-yl]-1,3-thiazol-2-yl]-2-methyl-propanamide | | Authors: | Wang, F, Cheng, W, Yuan, Z, Lin, D, Pan, W. | | Deposit date: | 2023-10-17 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Crystal Structure of LIMK2a from Biortus.
To Be Published
|
|
8X2S
 
 | | The Crystal Structure of BPGM from Biortus | | Descriptor: | 1,2-ETHANEDIOL, Bisphosphoglycerate mutase | | Authors: | Wang, F, Cheng, W, Yuan, Z, Qi, J, Li, J. | | Deposit date: | 2023-11-10 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Crystal Structure of BPGM from Biortus
To Be Published
|
|
8X2R
 
 | | The Crystal Structure of HSP 90-alpha from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Heat shock protein HSP 90-alpha | | Authors: | Wang, F, Cheng, W, Lv, Z, Meng, Q, Lu, Y. | | Deposit date: | 2023-11-10 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Crystal Structure of HSP 90-alpha from Biortus.
To Be Published
|
|
8WUG
 
 | | The Crystal Structure of JMJD2D from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Lysine-specific demethylase 4D, ... | | Authors: | Wang, F, Cheng, W, Yuan, Z, Lin, D, Guo, S. | | Deposit date: | 2023-10-20 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of JMJD2D from Biortus.
To Be Published
|
|
8X2P
 
 | | The Crystal Structure of LCK from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Wang, F, Cheng, W, Lv, Z, Meng, Q, Lu, Y. | | Deposit date: | 2023-11-10 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Crystal Structure of LCK from Biortus.
To Be Published
|
|
8X87
 
 | |
