4TTL
 
 | |
4Y33
 
 | | Crystal of NO66 in complex with Ni(II)and N-oxalylglycine (NOG) | | Descriptor: | Bifunctional lysine-specific demethylase and histidyl-hydroxylase NO66, N-OXALYLGLYCINE, NICKEL (II) ION | | Authors: | Wang, C, Zhang, Q, Zang, J. | | Deposit date: | 2015-02-10 | | Release date: | 2015-10-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the JmjC domain-containing protein NO66 complexed with ribosomal protein Rpl8.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1MHC
 
 | | MODEL OF MHC CLASS I H2-M3 WITH NONAPEPTIDE FROM RAT ND1 REFINED AT 2.3 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MHC CLASS I ANTIGEN H2-M3, NONAPEPTIDE FROM RAT NADH DEHYDROGENASE | | Authors: | Wang, C.-R, Fischer Lindahl, K, Deisenhofer, J. | | Deposit date: | 1995-08-23 | | Release date: | 1996-01-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Nonclassical binding of formylated peptide in crystal structure of the MHC class Ib molecule H2-M3
Cell(Cambridge,Mass.), 82, 1995
|
|
4TTO
 
 | |
4TTM
 
 | | Racemic structure of kalata B1 (kB1) | | Descriptor: | D-kalata B1, Kalata-B1 | | Authors: | Wang, C.K, King, G.J, Craik, D.J. | | Deposit date: | 2014-06-22 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9001 Å) | | Cite: | Racemic and Quasi-Racemic X-ray Structures of Cyclic Disulfide-Rich Peptide Drug Scaffolds.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4TTN
 
 | | Quasi-racemic structure of [G6A]kalata B1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, D-kalata B1, Kalata-B1 | | Authors: | Wang, C.K, King, G.J, Craik, D.J. | | Deposit date: | 2014-06-22 | | Release date: | 2014-09-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.2507 Å) | | Cite: | Racemic and Quasi-Racemic X-ray Structures of Cyclic Disulfide-Rich Peptide Drug Scaffolds.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4M36
 
 | |
6LPV
 
 | |
7XDG
 
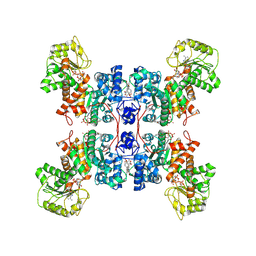 | | Cryo-EM structures of human mitochondrial NAD(P)+-dependent malic enzyme in a ternary complex with NAD+ and allosteric inhibitor MDSA | | Descriptor: | 5-[(3-carboxy-4-oxidanyl-phenyl)methyl]-2-oxidanyl-benzoic acid, NAD-dependent malic enzyme, mitochondrial, ... | | Authors: | Wang, C.H, Hsieh, J.T, Ho, M.C, Hung, H.C. | | Deposit date: | 2022-03-27 | | Release date: | 2023-03-29 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Suppression of the human malic enzyme 2 modifies energy metabolism and inhibits cellular respiration
Commun Biol, 6, 2023
|
|
7XDF
 
 | | Cryo-EM structures of human mitochondrial NAD(P)+-dependent malic enzyme in a ternary complex with NAD+ and allosteric inhibitor EA | | Descriptor: | 4-[(3-carboxy-2-oxidanyl-naphthalen-1-yl)methyl]-3-oxidanyl-naphthalene-2-carboxylic acid, NAD-dependent malic enzyme, mitochondrial, ... | | Authors: | Wang, C.H, Hsieh, J.T, Ho, M.C, Hung, H.C. | | Deposit date: | 2022-03-27 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Suppression of the human malic enzyme 2 modifies energy metabolism and inhibits cellular respiration
Commun Biol, 6, 2023
|
|
7XDE
 
 | | Cryo-EM structures of human mitochondrial NAD(P)+-dependent malic enzyme in apo form | | Descriptor: | NAD-dependent malic enzyme, mitochondrial, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Wang, C.H, Hsieh, J.T, Ho, M.C, Hung, H.C. | | Deposit date: | 2022-03-26 | | Release date: | 2023-03-29 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Suppression of the human malic enzyme 2 modifies energy metabolism and inhibits cellular respiration
Commun Biol, 6, 2023
|
|
8T0S
 
 | | Crystal structure of UBE2G2 adduct with phenethyl isothiocyanate (PEITC) at the Cys48 position | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase AMFR, Ubiquitin-conjugating enzyme E2 G2 | | Authors: | Wang, C, Shaw, G.X, Shi, G, Ji, X. | | Deposit date: | 2023-06-01 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of UBE2G2 adduct with phenethyl isothiocyanate (PEITC) at the Cys48 position
To be published
|
|
4M38
 
 | |
8DMF
 
 | | Cryo-EM structure of the ribosome-bound Bacteroides thetaiotaomicron EF-G2 | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Tetracycline resistance protein TetQ | | Authors: | Wang, C, Han, W, Groisman, E.A, Liu, J. | | Deposit date: | 2022-07-08 | | Release date: | 2023-01-04 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Gut colonization by Bacteroides requires translation by an EF-G paralog lacking GTPase activity.
Embo J., 2022
|
|
8EAN
 
 | |
8E4G
 
 | |
4M37
 
 | |
8DSP
 
 | |
8EAP
 
 | |
8EAO
 
 | |
1P5S
 
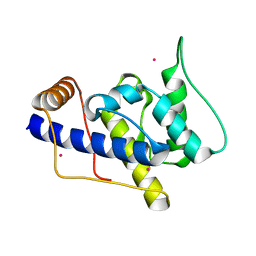 | | STRUCTURE AND FUNCTION OF THE CALPONIN-HOMOLOGY DOMAIN OF AN IQGAP PROTEIN FROM SCHIZOSACCHAROMYCES POMBE | | Descriptor: | MERCURY (II) ION, Ras GTPase-activating-like protein rng2 | | Authors: | Wang, C.H, Balasubramanian, M.K, Dokland, T. | | Deposit date: | 2003-04-28 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structure, crystal packing and molecular dynamics of the calponin-homology domain of Schizosaccharomyces pombe Rng2.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1P2X
 
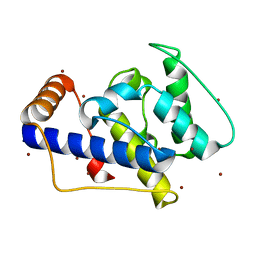 | | CRYSTAL STRUCTURE OF THE CALPONIN-HOMOLOGY DOMAIN OF RNG2 FROM SCHIZOSACCHAROMYCES POMBE | | Descriptor: | BROMIDE ION, Ras GTPase-activating-like protein | | Authors: | Wang, C.-H, Balasubramanian, M.K, Dokland, T. | | Deposit date: | 2003-04-16 | | Release date: | 2004-06-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure, crystal packing and molecular dynamics of the calponin-homology domain of Schizosaccharomyces pombe Rng2.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
5BOE
 
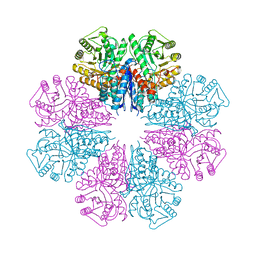 | | Crystal structure of Staphylococcus aureus enolase in complex with PEP | | Descriptor: | Enolase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Wang, C.L, Wu, Y.F, Han, L, Wu, M.H, Zhang, X, Zang, J.Y. | | Deposit date: | 2015-05-27 | | Release date: | 2015-12-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Octameric structure of Staphylococcus aureus enolase in complex with phosphoenolpyruvate
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7WN0
 
 | | Structure of PfENT1(Y190A) in complex with nanobody 19 | | Descriptor: | Equilibrative nucleoside/nucleobase transporter, nanobody19 | | Authors: | Wang, C, Deng, D, Ren, R.B, Yu, L.Y. | | Deposit date: | 2022-01-17 | | Release date: | 2023-02-01 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structural basis of the substrate recognition and inhibition mechanism of Plasmodium falciparum nucleoside transporter PfENT1.
Nat Commun, 14, 2023
|
|
8WX8
 
 | |
