2QD6
 
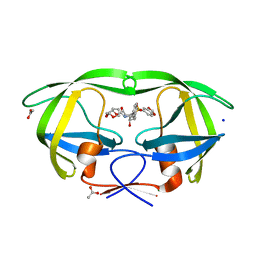 | | HIV-1 Protease Mutant I50V with potent Antiviral inhibitor GRL-98065 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(2S,3R)-3-HYDROXY-4-(N-ISOBUTYLBENZO[D][1,3]DIOXOLE-5-SULFONAMIDO)-1-PHENYLBUTAN-2-YLCARBAMATE, ACETATE ION, CHLORIDE ION, ... | | Authors: | Wang, Y.F, Tie, Y, Boross, P.I, Tozser, J, Ghosh, A.K, Harrison, R.W, Weber, I.T. | | Deposit date: | 2007-06-20 | | Release date: | 2008-04-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Potent new antiviral compound shows similar inhibition and structural interactions with drug resistant mutants and wild type HIV-1 protease.
J.Med.Chem., 50, 2007
|
|
5DMK
 
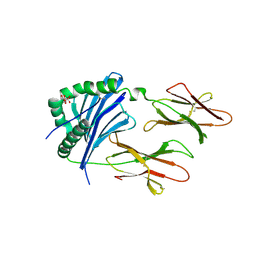 | | Crystal Structure of IAg7 in complex with RLGL-WE14 | | Descriptor: | CITRATE ANION, H-2 class II histocompatibility antigen, A-D alpha chain, ... | | Authors: | Wang, Y, Jin, N, Dai, S, Kappler, J.W. | | Deposit date: | 2015-09-08 | | Release date: | 2015-10-28 | | Last modified: | 2016-04-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | N-terminal additions to the WE14 peptide of chromogranin A create strong autoantigen agonists in type 1 diabetes.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5ZQ4
 
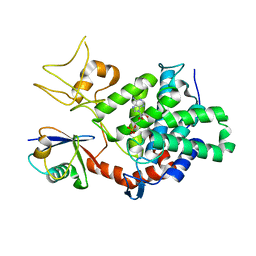 | | PDE-Ubi-ADPr | | Descriptor: | ADENOSINE MONOPHOSPHATE, SidE, ubiquitin | | Authors: | Wang, Y, Gao, A, Gao, P. | | Deposit date: | 2018-04-17 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.217 Å) | | Cite: | Structural Insights into Non-canonical Ubiquitination Catalyzed by SidE.
Cell, 173, 2018
|
|
4PFC
 
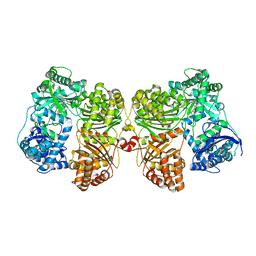 | | Crystal structure of insulin degrading enzyme complexed with inhibitor | | Descriptor: | Insulin-degrading enzyme, ZINC ION, methyl [(2S)-2-(5-{5-[4-({(2S)-2-[(3S)-3-amino-2-oxopiperidin-1-yl]-2-cyclohexylacetyl}amino)phenyl]pentyl}-2-fluorophenyl)-3-(quinolin-3-yl)propyl]carbamate | | Authors: | Wang, Y, Guo, S. | | Deposit date: | 2014-04-28 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Dual Exosite-binding Inhibitors of Insulin-degrading Enzyme Challenge Its Role as the Primary Mediator of Insulin Clearance in Vivo.
J.Biol.Chem., 290, 2015
|
|
5ZQ2
 
 | | SidE apo form | | Descriptor: | SidE | | Authors: | Wang, Y, Gao, A, Gao, P. | | Deposit date: | 2018-04-17 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | Structural Insights into Non-canonical Ubiquitination Catalyzed by SidE.
Cell, 173, 2018
|
|
3OK9
 
 | | Crystal structure of wild-type HIV-1 protease with new oxatricyclic designed inhibitor GRL-0519A | | Descriptor: | (3R,3aS,3bR,6aS,7aS)-octahydrodifuro[2,3-b:3',2'-d]furan-3-yl [(1S,2R)-1-benzyl-2-hydroxy-3-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}propyl]carbamate, CHLORIDE ION, GLYCEROL, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2010-08-24 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Probing Multidrug-Resistance and Protein-Ligand Interactions with Oxatricyclic Designed Ligands in HIV-1 Protease Inhibitors.
Chemmedchem, 5, 2010
|
|
7CAM
 
 | | SARS-CoV-2 main protease (Mpro) apo structure (space group P212121) | | Descriptor: | 3C-like proteinase | | Authors: | Wang, Y.C, Yang, C.S, Hou, M.H, Tsai, C.L, Chou, Y.Z, Chen, Y. | | Deposit date: | 2020-06-09 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis of SARS-CoV-2 main protease inhibition by a broad-spectrum anti-coronaviral drug.
Am J Cancer Res, 10, 2020
|
|
3HK2
 
 | | Crystal structure of T. thermophilus Argonaute N478 mutant protein complexed with DNA guide strand and 19-nt RNA target strand | | Descriptor: | 5'-D(P*TP*GP*AP*GP*GP*TP*AP*GP*TP*AP*GP*GP*TP*TP*GP*TP*AP*TP*AP*GP*T)-3', 5'-R(*UP*AP*UP*AP*CP*AP*AP*CP*CP*UP*AP*CP*UP*AP*CP*CP*UP*CP*G)-3', Argonaute, ... | | Authors: | Wang, Y, Li, H, Sheng, G, Patel, D.J. | | Deposit date: | 2009-05-22 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes.
Nature, 461, 2009
|
|
4PF9
 
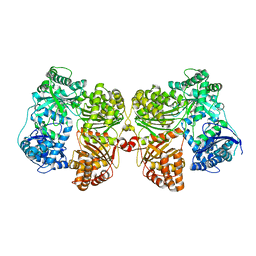 | | Crystal structure of insulin degrading enzyme complexed with inhibitor | | Descriptor: | Insulin-degrading enzyme, ZINC ION, methyl [(2S)-2-[4-({5-[4-({(2S)-2-[(3S)-3-amino-2-oxopiperidin-1-yl]-2-cyclohexylacetyl}amino)phenyl]pentyl}oxy)phenyl]-3-(quinolin-3-yl)propyl]carbamate | | Authors: | Wang, Y, Guo, S. | | Deposit date: | 2014-04-28 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dual Exosite-binding Inhibitors of Insulin-degrading Enzyme Challenge Its Role as the Primary Mediator of Insulin Clearance in Vivo.
J.Biol.Chem., 290, 2015
|
|
5XK2
 
 | |
4PF7
 
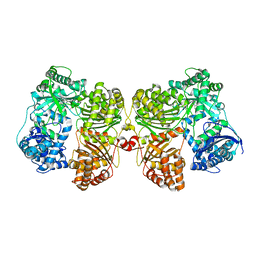 | | Crystal structure of insulin degrading enzyme complexed with inhibitor | | Descriptor: | (2S)-2-amino-N-{(1S)-1-cyclohexyl-2-[(4-methylphenyl)amino]-2-oxoethyl}-4-(methylselanyl)butanamide, Insulin-degrading enzyme, ZINC ION | | Authors: | Wang, Y, Guo, S. | | Deposit date: | 2014-04-28 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Dual Exosite-binding Inhibitors of Insulin-degrading Enzyme Challenge Its Role as the Primary Mediator of Insulin Clearance in Vivo.
J.Biol.Chem., 290, 2015
|
|
5ZQ3
 
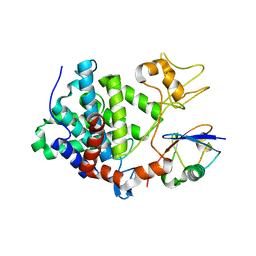 | | PDE-Ubiquitin | | Descriptor: | SidE, Ubiquitin | | Authors: | Wang, Y, Gao, A, Gao, P. | | Deposit date: | 2018-04-17 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.182 Å) | | Cite: | Structural Insights into Non-canonical Ubiquitination Catalyzed by SidE.
Cell, 173, 2018
|
|
5ZQ5
 
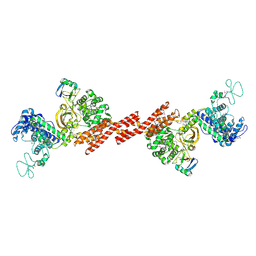 | | SidE-Ubi | | Descriptor: | SidE, Ubiquitin | | Authors: | Wang, Y, Gao, A, Gao, P. | | Deposit date: | 2018-04-17 | | Release date: | 2018-05-23 | | Last modified: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (2.487 Å) | | Cite: | Structural Insights into Non-canonical Ubiquitination Catalyzed by SidE.
Cell, 173, 2018
|
|
1R6N
 
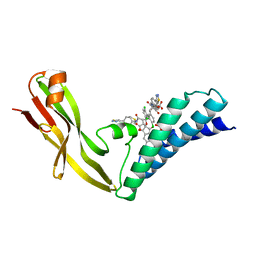 | | HPV11 E2 TAD complex crystal structure | | Descriptor: | 2-METHYL-PROPIONIC ACID, DIMETHYL SULFOXIDE, HPV11 REGULATORY PROTEIN E2, ... | | Authors: | Wang, Y, Coulombe, R. | | Deposit date: | 2003-10-15 | | Release date: | 2004-02-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the E2 Transactivation Domain of Human
Papillomavirus Type 11 Bound to a Protein Interaction Inhibitor
J.Biol.Chem., 279, 2004
|
|
7E0V
 
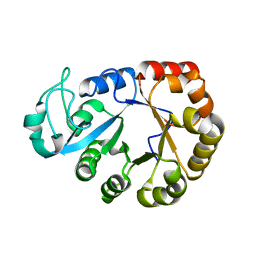 | | GDPD from Pyrococcus furiosus DSM 3638 | | Descriptor: | Glycerophosphodiester phosphodiesterase | | Authors: | Wang, Y.H, Wang, J, Wang, F.H. | | Deposit date: | 2021-01-28 | | Release date: | 2022-02-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of Bomgl, a monoacylglycerol lipase from marine Bacillus
To Be Published
|
|
3QAA
 
 | | HIV-1 wild type protease with a substituted bis-Tetrahydrofuran inhibitor, GRL-044-10A | | Descriptor: | (3R,3aS,4R,6aR)-4-methoxyhexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, GLYCEROL, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2011-01-10 | | Release date: | 2011-12-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Design of substituted bis-Tetrahydrofuran (bis-THF)-derived Potent HIV-1 Protease Inhibitors, Protein-ligand X-ray Structure, and Convenient Syntheses of bis-THF and Substituted bis-THF Ligands.
ACS Med Chem Lett, 2, 2011
|
|
3HVR
 
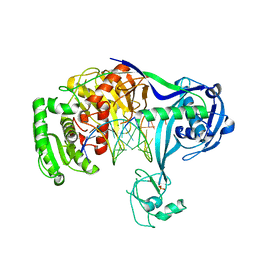 | | Crystal structure of T. thermophilus Argonaute complexed with DNA guide strand and 19-nt RNA target strand with two Mg2+ at the cleavage site | | Descriptor: | 5'-D(P*TP*GP*AP*GP*GP*TP*AP*GP*TP*AP*GP*GP*TP*TP*TP*GP*AP*TP*AP*GP*T)-3', 5'-R(*UP*AP*UP*AP*CP*AP*AP*CP*CP*UP*AP*CP*UP*AP*CP*CP*UP*CP*G)-3', Argonaute, ... | | Authors: | Wang, Y, Li, H, Sheng, G, Patel, D.J. | | Deposit date: | 2009-06-16 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.211 Å) | | Cite: | Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes.
Nature, 461, 2009
|
|
5G4C
 
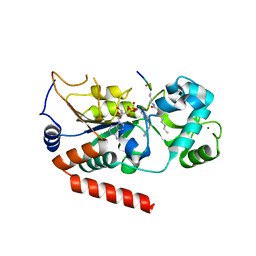 | | Human SIRT2 catalyse short chain fatty acyl lysine | | Descriptor: | CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, NAD-DEPENDENT PROTEIN DEACETYLASE SIRTUIN-2, SIRT2, ... | | Authors: | Wang, Y. | | Deposit date: | 2016-05-09 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | SIRT2 Reverses 4-Oxononanoyl Lysine Modification on Histones.
J. Am. Chem. Soc., 138, 2016
|
|
7F9Y
 
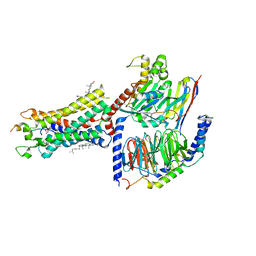 | | ghrelin-bound ghrelin receptor in complex with Gq | | Descriptor: | CHOLESTEROL, Engineered G-alpha-q subunit, Ghrelin-28, ... | | Authors: | Wang, Y, Zhuang, Y, Xu, P, Xu, H.E, Jiang, Y. | | Deposit date: | 2021-07-05 | | Release date: | 2021-08-18 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular recognition of an acyl-peptide hormone and activation of ghrelin receptor.
Nat Commun, 12, 2021
|
|
5H00
 
 | |
5ZQ7
 
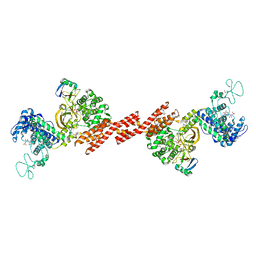 | | SidE-Ubi-NAD | | Descriptor: | ADENOSINE MONOPHOSPHATE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SidE, ... | | Authors: | Wang, Y, Gao, A, Gao, P. | | Deposit date: | 2018-04-17 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.847 Å) | | Cite: | Structural Insights into Non-canonical Ubiquitination Catalyzed by SidE.
Cell, 173, 2018
|
|
5GZX
 
 | |
5ZQ6
 
 | | SidE-Ubi-ADPr | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, SidE, Ubiquitin | | Authors: | Wang, Y, Gao, A, Gao, P. | | Deposit date: | 2018-04-17 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.009 Å) | | Cite: | Structural Insights into Non-canonical Ubiquitination Catalyzed by SidE.
Cell, 173, 2018
|
|
7KQR
 
 | | A 1.89-A resolution substrate-bound crystal structure of heme-dependent tyrosine hydroxylase from S. sclerotialus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Heme-dependent L-tyrosine hydroxylase, ... | | Authors: | Wang, Y, Shin, I, Liu, A. | | Deposit date: | 2020-11-17 | | Release date: | 2021-03-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Molecular Rationale for Partitioning between C-H and C-F Bond Activation in Heme-Dependent Tyrosine Hydroxylase.
J.Am.Chem.Soc., 143, 2021
|
|
7KQS
 
 | | A 1.68-A resolution 3-fluoro-L-tyrosine bound crystal structure of heme-dependent tyrosine hydroxylase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-FLUOROTYROSINE, ... | | Authors: | Wang, Y, Liu, A. | | Deposit date: | 2020-11-17 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.677 Å) | | Cite: | Molecular Rationale for Partitioning between C-H and C-F Bond Activation in Heme-Dependent Tyrosine Hydroxylase.
J.Am.Chem.Soc., 143, 2021
|
|
