1AIH
 
 | | CATALYTIC DOMAIN OF BACTERIOPHAGE HP1 INTEGRASE | | Descriptor: | HP1 INTEGRASE, MAGNESIUM ION, SULFATE ION | | Authors: | Hickman, A.B, Waninger, S, Scocca, J.J, Dyda, F. | | Deposit date: | 1997-04-17 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular organization in site-specific recombination: the catalytic domain of bacteriophage HP1 integrase at 2.7 A resolution.
Cell(Cambridge,Mass.), 89, 1997
|
|
3NO9
 
 | | Crystal Structure of apo fumarate hydratase from Mycobacterium tuberculosis | | Descriptor: | Fumarate hydratase class II | | Authors: | Li, H, Swanson, S, Yu, M, Hung, L.-W, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-06-25 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal Structure of apo fumarate hydratase from Mycobacterium tuberculosis
To be Published
|
|
1CXV
 
 | | STRUCTURE OF RECOMBINANT MOUSE COLLAGENASE-3 (MMP-13) | | Descriptor: | 2-{4-[4-(4-CHLORO-PHENOXY)-BENZENESULFONYL]-TETRAHYDRO-PYRAN-4-YL}-N-HYDROXY-ACETAMIDE, CALCIUM ION, PROTEIN (COLLAGENASE-3), ... | | Authors: | Botos, I, Meyer, E, Swanson, S.M, Lemaitre, V, Eeckhout, Y, Meyer, E.F. | | Deposit date: | 1999-08-30 | | Release date: | 2000-08-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of recombinant mouse collagenase-3 (MMP-13).
J.Mol.Biol., 292, 1999
|
|
1WNI
 
 | | Crystal Structure of H2-Proteinase | | Descriptor: | Trimerelysin II, ZINC ION | | Authors: | Kumasaka, T, Yamamoto, M, Moriyama, H, Tanaka, N, Sato, M, Katsube, Y, Yamakawa, Y, Omori-Satoh, T, Iwanaga, S, Ueki, T. | | Deposit date: | 2004-08-04 | | Release date: | 2004-08-17 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of H2-proteinase from the venom of Trimeresurus flavoviridis.
J.Biochem., 119, 1996
|
|
7DTL
 
 | | Crystal structure of PSK, an antimicrobial peptide from Chrysomya megacephala | | Descriptor: | PSK | | Authors: | Xiao, C, Xiao, Z, Wang, S, Liu, W. | | Deposit date: | 2021-01-05 | | Release date: | 2021-07-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal and solution structures of a novel antimicrobial peptide from Chrysomya megacephala.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
1WZZ
 
 | | Structure of endo-beta-1,4-glucanase CMCax from Acetobacter xylinum | | Descriptor: | Probable endoglucanase, SULFATE ION | | Authors: | Yasutake, Y, Kawano, S, Tajima, K, Yao, M, Satoh, Y, Munekata, M, Tanaka, I, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-03-10 | | Release date: | 2006-03-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural characterization of the Acetobacter xylinum endo-beta-1,4-glucanase CMCax required for cellulose biosynthesis.
Proteins, 64, 2006
|
|
2RQ8
 
 | | Solution NMR structure of titin I27 domain mutant | | Descriptor: | Titin | | Authors: | Yagawa, K, Oguro, T, Momose, T, Kawano, S, Sato, T, Endo, T. | | Deposit date: | 2009-03-05 | | Release date: | 2010-02-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for unfolding pathway-dependent stability of proteins: Vectorial unfolding vs. global unfolding
Protein Sci., 2010
|
|
2MUQ
 
 | | Solution Structure of the Human FAAP20 UBZ | | Descriptor: | Fanconi anemia-associated protein of 20 kDa, ZINC ION | | Authors: | Wojtaszek, J.L, Wang, S, Zhou, P. | | Deposit date: | 2014-09-16 | | Release date: | 2014-12-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ubiquitin recognition by FAAP20 expands the complex interface beyond the canonical UBZ domain.
Nucleic Acids Res., 42, 2014
|
|
2VIW
 
 | | Fragment-Based Discovery of Mexiletine Derivatives as Orally Bioavailable Inhibitors of Urokinase-Type Plasminogen Activator | | Descriptor: | 4-(2-aminoethoxy)-N-(3-chloro-2-ethoxy-5-piperidin-1-ylphenyl)-3,5-dimethylbenzamide, ACETATE ION, UROKINASE-TYPE PLASMINOGEN ACTIVATOR CHAIN B | | Authors: | Frederickson, M, Callaghan, O, Chessari, G, Congreve, M, Cowan, S.R, Matthews, J.E, McMenamin, R, Smith, D, Vinkovic, M, Wallis, N.G. | | Deposit date: | 2007-12-05 | | Release date: | 2008-01-22 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Fragment-Based Discovery of Mexiletine Derivatives as Orally Bioavailable Inhibitors of Urokinase-Type Plasminogen Activator.
J.Med.Chem., 51, 2008
|
|
2VIV
 
 | | Fragment-Based Discovery of Mexiletine Derivatives as Orally Bioavailable Inhibitors of Urokinase-Type Plasminogen Activator | | Descriptor: | 4-(2-aminoethoxy)-N-(3-chloro-5-piperidin-1-ylphenyl)-3,5-dimethylbenzamide, ACETATE ION, UROKINASE-TYPE PLASMINOGEN ACTIVATOR CHAIN B | | Authors: | Frederickson, M, Callaghan, O, Chessari, G, Congreve, M, Cowan, S.R, Matthews, J.E, McMenamin, R, Smith, D, Vinkovic, M, Wallis, N.G. | | Deposit date: | 2007-12-05 | | Release date: | 2008-01-22 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Fragment-Based Discovery of Mexiletine Derivatives as Orally Bioavailable Inhibitors of Urokinase-Type Plasminogen Activator.
J.Med.Chem., 51, 2008
|
|
2VIQ
 
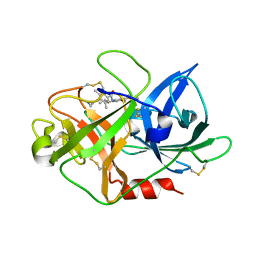 | | Fragment-Based Discovery of Mexiletine Derivatives as Orally Bioavailable Inhibitors of Urokinase-Type Plasminogen Activator | | Descriptor: | 4-(2-aminoethoxy)-N-(2,5-diethoxyphenyl)-3,5-dimethylbenzamide, ACETATE ION, UROKINASE-TYPE PLASMINOGEN ACTIVATOR CHAIN B | | Authors: | Frederickson, M, Callaghan, O, Chessari, G, Congreve, M, Cowan, S.R, Matthews, J.E, McMenamin, R, Smith, D, Vinkovic, M, Wallis, N.G. | | Deposit date: | 2007-12-05 | | Release date: | 2008-01-22 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fragment-Based Discovery of Mexiletine Derivatives as Orally Bioavailable Inhibitors of Urokinase-Type Plasminogen Activator.
J.Med.Chem., 51, 2008
|
|
2VIO
 
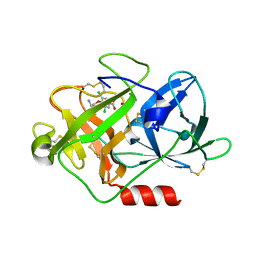 | | Fragment-Based Discovery of Mexiletine Derivatives as Orally Bioavailable Inhibitors of Urokinase-Type Plasminogen Activator | | Descriptor: | 4-(2-aminoethoxy)-3,5-dichlorobenzoic acid, ACETATE ION, UROKINASE-TYPE PLASMINOGEN ACTIVATOR CHAIN B | | Authors: | Frederickson, M, Callaghan, O, Chessari, G, Congreve, M, Cowan, S.R, Matthews, J.E, McMenamin, R, Smith, D, Vinkovic, M, Wallis, N.G. | | Deposit date: | 2007-12-05 | | Release date: | 2008-01-22 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fragment-Based Discovery of Mexiletine Derivatives as Orally Bioavailable Inhibitors of Urokinase-Type Plasminogen Activator
J.Med.Chem., 51, 2008
|
|
2O5U
 
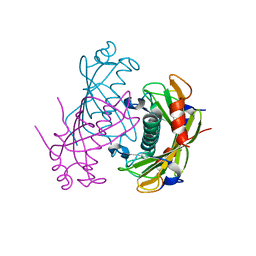 | | Crystal structure of the PA5185 protein from Pseudomonas Aeruginosa strain PAO1- orthorhombic form (C222). | | Descriptor: | Thioesterase | | Authors: | Chruszcz, M, Wang, S, Evdokimova, E, Koclega, K.D, Kudritska, M, Savchenko, A, Edwards, A, Minor, W. | | Deposit date: | 2006-12-06 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Function-biased choice of additives for optimization of protein crystallization - the case of the putative thioesterase PA5185 from Pseudomonas aeruginosa PAO1.
Cryst.Growth Des., 8, 2008
|
|
3RTO
 
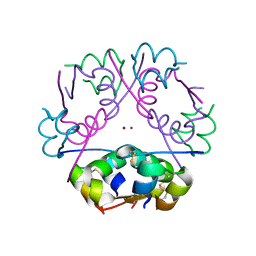 | | Acoustically mounted porcine insulin microcrystals yield an X-ray SAD structure | | Descriptor: | Insulin, ZINC ION | | Authors: | Soares, A.S, Engel, M.A, Stearns, R, Datwani, S, Olechno, J, Ellson, R, Skinner, J.M, Allaire, M, Orville, A.M. | | Deposit date: | 2011-05-03 | | Release date: | 2011-05-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Acoustically Mounted Microcrystals Yield High-Resolution X-ray Structures.
Biochemistry, 50, 2011
|
|
2O0Y
 
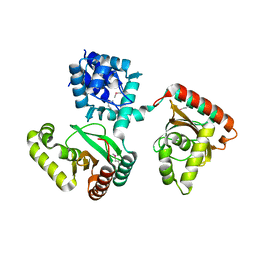 | | Crystal structure of putative transcriptional regulator RHA1_ro06953 (IclR-family) from Rhodococcus sp. | | Descriptor: | Transcriptional regulator | | Authors: | Chruszcz, M, Wang, S, Skarina, T, Onopriyenko, O, Cymborowski, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-11-28 | | Release date: | 2006-12-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Putative Transcriptional Regul 2 Rha1_Ro06953(Iclr-Family) from Rhodococcus Sp.
To be Published
|
|
3AAU
 
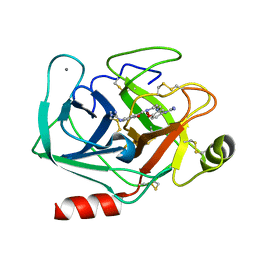 | | Bovine beta-trypsin bound to meta-diguanidino schiff base copper (II) chelate | | Descriptor: | CALCIUM ION, COPPER (II) ION, Cationic trypsin, ... | | Authors: | Iyaguchi, D, Kawano, S, Toyota, E. | | Deposit date: | 2009-11-26 | | Release date: | 2010-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the design of novel Schiff base metal chelate inhibitors of trypsin
Bioorg.Med.Chem., 18, 2010
|
|
3AAS
 
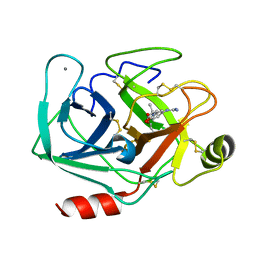 | | Bovine beta-trypsin bound to meta-guanidino schiff base copper (II) chelate | | Descriptor: | (E)-N-[(5-carbamimidamido-2-hydroxyphenyl)methylidene]-L-alanine, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Iyaguchi, D, Kawano, S, Toyota, E. | | Deposit date: | 2009-11-26 | | Release date: | 2010-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for the design of novel Schiff base metal chelate inhibitors of trypsin
Bioorg.Med.Chem., 18, 2010
|
|
2DUT
 
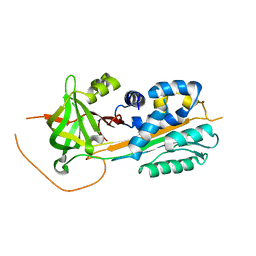 | |
7WVH
 
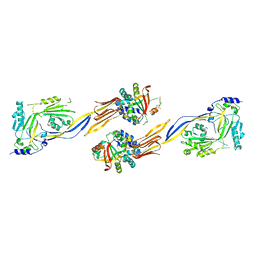 | |
2LWG
 
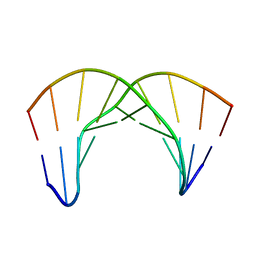 | |
2LWH
 
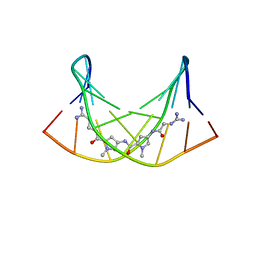 | | NMR Structure of the Self-Complementary 10 mer DNA Duplex 5'-GGATATATCC-3' in Complex with Netropsin | | Descriptor: | DNA (5'-D(*GP*GP*AP*TP*AP*TP*AP*TP*CP*C)-3'), NETROPSIN | | Authors: | Rettig, M, Germann, M.W, Wilson, W, Wang, S. | | Deposit date: | 2012-07-31 | | Release date: | 2013-01-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for sequence-dependent induced DNA bending.
Chembiochem, 14, 2013
|
|
1AOC
 
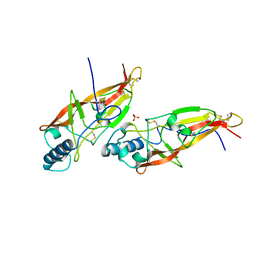 | | JAPANESE HORSESHOE CRAB COAGULOGEN | | Descriptor: | COAGULOGEN, SULFATE ION | | Authors: | Bergner, A, Oganessyan, V, Muta, T, Iwanaga, S, Typke, D, Huber, R, Bode, W. | | Deposit date: | 1996-11-28 | | Release date: | 1997-04-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a coagulogen, the clotting protein from horseshoe crab: a structural homologue of nerve growth factor.
EMBO J., 15, 1996
|
|
2H4R
 
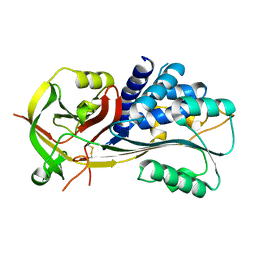 | |
5E5V
 
 | | Structure of amyloid forming peptide NFGAILS (residues 22-28) from Islet Amyloid Polypeptide | | Descriptor: | NFGAILS (22-28) from islet amyloid polypeptide, synthesized | | Authors: | Soriaga, A.B, Macdonald, R, Sawaya, M.R, Sangwan, S, Eisenberg, D. | | Deposit date: | 2015-10-09 | | Release date: | 2015-12-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Crystal Structures of IAPP Amyloidogenic Segments Reveal a Novel Packing Motif of Out-of-Register Beta Sheets.
J.Phys.Chem.B, 120, 2016
|
|
3AAV
 
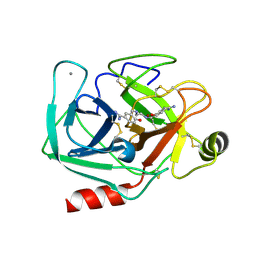 | | Bovine beta-trypsin bound to meta-diamidino schiff base copper (II) chelate | | Descriptor: | 3,3'-[ethane-1,2-diylbis(nitrilomethylylidene)]bis(4-hydroxybenzenecarboximidamide), CALCIUM ION, COPPER (II) ION, ... | | Authors: | Iyaguchi, D, Kawano, S, Toyota, E. | | Deposit date: | 2009-11-26 | | Release date: | 2010-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the design of novel Schiff base metal chelate inhibitors of trypsin
Bioorg.Med.Chem., 18, 2010
|
|
