7SQD
 
 | |
8FXR
 
 | | Structure of neck with portal vertex of capsid of Agrobacterium phage Milano | | Descriptor: | Collar sheath protein, gp13, Linking protein 1, ... | | Authors: | Sonani, R.R, Wang, F, Esteves, N.C, Kelly, R.J, Sebastian, A, Kreutzberger, M.A.B, Leiman, P.G, Scharf, B.E, Egelman, E.H. | | Deposit date: | 2023-01-25 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Neck and capsid architecture of the robust Agrobacterium phage Milano.
Commun Biol, 6, 2023
|
|
8FWM
 
 | | Structure of tail-neck junction of Agrobacterium phage Milano | | Descriptor: | Collar sheath protein, gp13, Tail sheath protein, ... | | Authors: | Sonani, R.R, Wang, F, Esteves, N.C, Kelly, R.J, Sebastian, A, Kreutzberger, M.A.B, Leiman, P.G, Scharf, B.E, Egelman, E.H. | | Deposit date: | 2023-01-23 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Neck and capsid architecture of the robust Agrobacterium phage Milano.
Commun Biol, 6, 2023
|
|
8WUL
 
 | | Crystal structure of affinity enhanced TCR in complex with HLA-A*11:01 bound to KRAS-G12V peptide (VVGAVGVGK) | | Descriptor: | Beta-2-microglobulin, KRAS-G12V nonamer peptide, MHC class I antigen, ... | | Authors: | Zhang, M.Y, Luo, L.J, Xu, W, Guan, F.H, Wang, X.Y, Zhu, P, Zhang, J.H, Zhou, X.Y, Wang, F, Ye, S. | | Deposit date: | 2023-10-20 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Identification and affinity enhancement of T-cell receptor targeting a KRAS G12V cancer neoantigen.
Commun Biol, 7, 2024
|
|
8FQC
 
 | | Structure of baseplate with receptor binding complex of Agrobacterium phage Milano | | Descriptor: | Baseplate Centerpiece, gp25, Baseplate Central Spike, ... | | Authors: | Sonani, R.R, Leiman, P.G, Wang, F, Kreutzberger, M.A.B, Sebastian, A, Esteves, N.C, Kelly, R.J, Scharf, B, Egelman, E.H. | | Deposit date: | 2023-01-05 | | Release date: | 2024-01-31 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | An extensive disulfide bond network prevents tail contraction in Agrobacterium tumefaciens phage Milano.
Nat Commun, 15, 2024
|
|
8WTE
 
 | | Crystal structure of TCR in complex with HLA-A*11:01 bound to KRAS-G12V peptide (VVGAVGVGK) | | Descriptor: | Beta-2-microglobulin, KRAS-G12V nonamer peptide, MHC class I antigen (Fragment), ... | | Authors: | Zhang, M.Y, Luo, L.J, Xu, W, Guan, F.H, Wang, X.Y, Zhu, P, Zhang, J.H, Zhou, X.Y, Wang, F, Ye, S. | | Deposit date: | 2023-10-18 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Identification and affinity enhancement of T-cell receptor targeting a KRAS G12V cancer neoantigen.
Commun Biol, 7, 2024
|
|
3RLR
 
 | | Co-crystal structure of the HSP90 ATP binding domain in complex with 4-(2,4-dichloro-5-methoxyphenyl)-2,6-dimethyl-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile | | Descriptor: | 4-(2,4-dichloro-5-methoxyphenyl)-2,6-dimethyl-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, Heat shock protein HSP 90-alpha, PHOSPHATE ION | | Authors: | Kung, P.-P, Sinnema, P.-J, Richardson, P, Hickey, M.J, Gajiwala, K.S, Wang, F, Huang, B, McClellan, G, Wang, J, Maegley, K, Bergqvist, S, Mehta, P.P, Kania, R. | | Deposit date: | 2011-04-20 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design strategies to target crystallographic waters applied to the Hsp90 molecular chaperone.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3RLQ
 
 | | Co-crystal structure of the HSP90 ATP binding domain in complex with 4-(2,4-dichloro-5-methoxyphenyl)-2-methyl-7H-pyrrolo[2,3-d]pyrimidine-5- carbonitrile | | Descriptor: | 4-(2,4-dichloro-5-methoxyphenyl)-2-methyl-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, Heat shock protein HSP 90-alpha, PHOSPHATE ION | | Authors: | Kung, P.-P, Sinnema, P.-J, Richardson, P, Hickey, M.J, Gajiwala, K.S, Wang, F, Huang, B, McClellan, G, Wang, J, Maegley, K, Bergqvist, S, Mehta, P.P, Kania, R. | | Deposit date: | 2011-04-20 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design strategies to target crystallographic waters applied to the Hsp90 molecular chaperone.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3JCP
 
 | | Structure of yeast 26S proteasome in M2 state derived from Titan dataset | | Descriptor: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | Authors: | Luan, B, Huang, X.L, Wu, J.P, Shi, Y.G, Wang, F. | | Deposit date: | 2016-01-06 | | Release date: | 2016-06-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structure of an endogenous yeast 26S proteasome reveals two major conformational states.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3RLP
 
 | | Co-crystal structure of the HSP90 ATP binding domain in complex with 4-(2,4-dichloro-5-methoxyphenyl)-6-methylpyrimidin-2-amine | | Descriptor: | 4-(2,4-dichloro-5-methoxyphenyl)-6-methylpyrimidin-2-amine, Heat shock protein HSP 90-alpha, PHOSPHATE ION | | Authors: | Kung, P.-P, Sinnema, P.-J, Richardson, P, Hickey, M.J, Gajiwala, K.S, Wang, F, Huang, B, McClellan, G, Wang, J, Maegley, K, Bergqvist, S, Mehta, P.P, Kania, R. | | Deposit date: | 2011-04-20 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design strategies to target crystallographic waters applied to the Hsp90 molecular chaperone.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3JCI
 
 | | 2.9 Angstrom Resolution Cryo-EM 3-D Reconstruction of Close-packed PCV2 Virus-like Particles | | Descriptor: | Capsid protein | | Authors: | Liu, Z, Guo, F, Wang, F, Li, T.C, Jiang, W. | | Deposit date: | 2015-12-13 | | Release date: | 2016-02-03 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | 2.9 angstrom Resolution Cryo-EM 3D Reconstruction of Close-Packed Virus Particles.
Structure, 24, 2016
|
|
4QY0
 
 | | Structure of H10 from human-infecting H10N8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, hemagglutinin | | Authors: | Wang, M, Zhang, W, Qi, J, Wang, F, Zhou, J, Bi, Y, Wu, Y, Sun, H, Liu, J, Huang, C, Li, X, Yan, J, Shu, Y, Shi, Y, Gao, G.F. | | Deposit date: | 2014-07-23 | | Release date: | 2015-01-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structural basis for preferential avian receptor binding by the human-infecting H10N8 avian influenza virus
Nat Commun, 6, 2015
|
|
4QY2
 
 | | Structure of H10 from human-infecting H10N8 virus in complex with human receptor analog | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-acetyl-alpha-neuraminic acid, hemagglutinin | | Authors: | Wang, M, Zhang, W, Qi, J, Wang, F, Zhou, J, Bi, Y, Wu, Y, Sun, H, Liu, J, Huang, C, Li, X, Yan, J, Shu, Y, Shi, Y, Gao, G.F. | | Deposit date: | 2014-07-23 | | Release date: | 2015-01-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Structural basis for preferential avian receptor binding by the human-infecting H10N8 avian influenza virus
Nat Commun, 6, 2015
|
|
4ZMI
 
 | | Crystal structure of the Helical domain of S. pombe Taz1 | | Descriptor: | COBALT (II) ION, MAGNESIUM ION, Telomere length regulator taz1 | | Authors: | Deng, W, Wu, J, Wang, F, Lei, M. | | Deposit date: | 2015-05-04 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fission yeast telomere-binding protein Taz1 is a functional but not a structural counterpart of human TRF1 and TRF2.
Cell Res., 25, 2015
|
|
4KN8
 
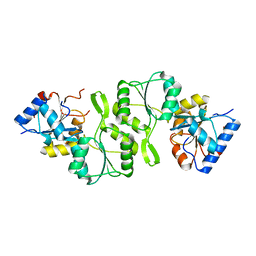 | | Crystal structure of Bs-TpNPPase | | Descriptor: | Thermostable NPPase | | Authors: | Guo, Z, Wang, F, Huang, J, Gong, W, Ji, C. | | Deposit date: | 2013-05-09 | | Release date: | 2014-04-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Crystal Structure of Thermostable p-nitrophenylphosphatase from Bacillus Stearothermophilus (Bs-TpNPPase)
PROTEIN PEPT.LETT., 21, 2014
|
|
6K9N
 
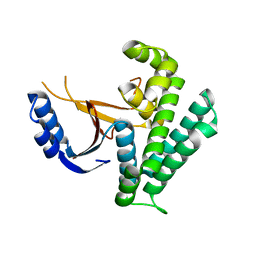 | | Rice_OTUB_like_catalytic domain | | Descriptor: | Ubiquitin thioesterase | | Authors: | Lu, L.N, Liu, L, Wang, F. | | Deposit date: | 2019-06-17 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Met1-specific motifs conserved in OTUB subfamily of green plants enable rice OTUB1 to hydrolyse Met1 ubiquitin chains
Nat Commun, 13, 2022
|
|
4ZWV
 
 | | Crystal Structure of Aminotransferase AtmS13 from Actinomadura melliaura | | Descriptor: | GLYCEROL, Putative aminotransferase | | Authors: | Kim, Y, Bigelow, L, Endres, M, Wang, F, Phillips Jr, G.N, Joachimiak, A, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-05-19 | | Release date: | 2015-06-03 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (1.503 Å) | | Cite: | Structural characterization of AtmS13, a putative sugar aminotransferase involved in indolocarbazole AT2433 aminopentose biosynthesis.
Proteins, 83, 2015
|
|
6IIL
 
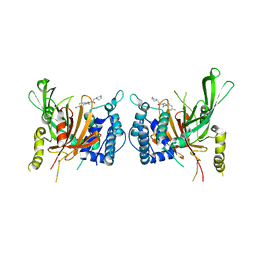 | | USP14 catalytic domain bind to IU1-47 | | Descriptor: | 1-[1-(4-chlorophenyl)-2,5-dimethyl-1H-pyrrol-3-yl]-2-(piperidin-1-yl)ethan-1-one, Ubiquitin carboxyl-terminal hydrolase 14 | | Authors: | Mei, Z.Q, Wang, Y.W, Wang, F, Wang, J.W, He, W, Ding, S, Li, J.W. | | Deposit date: | 2018-10-07 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Small molecule inhibitors reveal allosteric regulation of USP14 via steric blockade.
Cell Res., 28, 2018
|
|
6KBE
 
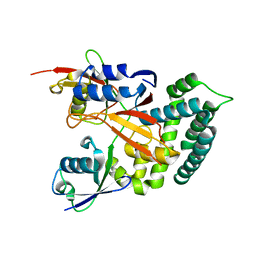 | | Structure of Deubiquitinase | | Descriptor: | Polyubiquitin-C, Ubiquitin thioesterase | | Authors: | Lu, L.N, Liu, L, Wang, F. | | Deposit date: | 2019-06-24 | | Release date: | 2020-06-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.339 Å) | | Cite: | Met1-specific motifs conserved in OTUB subfamily of green plants enable rice OTUB1 to hydrolyse Met1 ubiquitin chains
Nat Commun, 13, 2022
|
|
6K9P
 
 | | Structure of Deubiquitinase | | Descriptor: | Ubiquitin, Ubiquitin thioesterase | | Authors: | Lu, L.N, Liu, L, Wang, F. | | Deposit date: | 2019-06-17 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | Met1-specific motifs conserved in OTUB subfamily of green plants enable rice OTUB1 to hydrolyse Met1 ubiquitin chains
Nat Commun, 13, 2022
|
|
7SLZ
 
 | | CRYSTAL STRUCTURE OF GID4 IN COMPLEX WITH BPF023596 | | Descriptor: | Glucose-induced degradation protein 4 homolog, N-[(1s,4s)-4-(1H-benzimidazol-2-yl)cyclohexyl]-N~2~-[(1H-indol-2-yl)methyl]glycinamide | | Authors: | Song, X, Dong, A, Calabrese, M, Wang, F, Owen, D, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-10-25 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | CRYSTAL STRUCTURE OF GID4 IN COMPLEX WITH BPF023596
To Be Published
|
|
6IIM
 
 | | USP14 catalytic domain with IU1-206 | | Descriptor: | 1-[1-(4-chlorophenyl)-2,5-dimethyl-1H-pyrrol-3-yl]-2-(4-hydroxypiperidin-1-yl)ethan-1-one, Ubiquitin carboxyl-terminal hydrolase 14 | | Authors: | Mei, Z.Q, Wang, J.W, Wang, F, Wang, Y.W. | | Deposit date: | 2018-10-07 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Small molecule inhibitors reveal allosteric regulation of USP14 via steric blockade.
Cell Res., 28, 2018
|
|
6IIN
 
 | | USP14 catalytic domain with IU1-248 | | Descriptor: | 4-{3-[(4-hydroxypiperidin-1-yl)acetyl]-2,5-dimethyl-1H-pyrrol-1-yl}benzonitrile, Ubiquitin carboxyl-terminal hydrolase 14 | | Authors: | Mei, Z.Q, Wang, J.W, Wang, F, Wang, Y.W. | | Deposit date: | 2018-10-07 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Small molecule inhibitors reveal allosteric regulation of USP14 via steric blockade.
Cell Res., 28, 2018
|
|
4ZMK
 
 | |
3J3R
 
 | | Structural dynamics of the MecA-ClpC complex revealed by cryo-EM | | Descriptor: | Adapter protein MecA 1, Negative regulator of genetic competence ClpC/MecB | | Authors: | Liu, J, Mei, Z, Li, N, Qi, Y, Xu, Y, Shi, Y, Wang, F, Lei, J, Gao, N. | | Deposit date: | 2013-04-18 | | Release date: | 2013-05-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (9.4 Å) | | Cite: | Structural dynamics of the MecA-ClpC complex: a type II AAA+ protein unfolding machine
J.Biol.Chem., 288, 2013
|
|
