3LZT
 
 | | REFINEMENT OF TRICLINIC LYSOZYME AT ATOMIC RESOLUTION | | Descriptor: | ACETATE ION, LYSOZYME, NITRATE ION | | Authors: | Walsh, M.A, Schneider, T, Sieker, L.C, Dauter, Z, Lamzin, V, Wilson, K.S. | | Deposit date: | 1997-03-23 | | Release date: | 1998-03-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (0.925 Å) | | Cite: | Refinement of triclinic hen egg-white lysozyme at atomic resolution.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
4LZT
 
 | | ATOMIC RESOLUTION REFINEMENT OF TRICLINIC HEW LYSOZYME AT 295K | | Descriptor: | LYSOZYME, NITRATE ION | | Authors: | Walsh, M.A, Schneider, T, Sieker, L.C, Dauter, Z, Lamzin, V, Wilson, K.S. | | Deposit date: | 1997-03-31 | | Release date: | 1998-04-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Refinement of triclinic hen egg-white lysozyme at atomic resolution.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1BU5
 
 | | X-RAY CRYSTAL STRUCTURE OF THE DESULFOVIBRIO VULGARIS (HILDENBOROUGH) APOFLAVODOXIN-RIBOFLAVIN COMPLEX | | Descriptor: | PROTEIN (FLAVODOXIN), RIBOFLAVIN, SULFATE ION | | Authors: | Walsh, M.A, Mccarthy, A, O'Farrell, P.A, Mccardle, P, Cunningham, P.D, Mayhew, S.G, Higgins, T.M. | | Deposit date: | 1998-09-12 | | Release date: | 1999-02-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | X-ray crystal structure of the Desulfovibrio vulgaris (Hildenborough) apoflavodoxin-riboflavin complex.
Eur.J.Biochem., 258, 1998
|
|
1AZL
 
 | | G61V FLAVODOXIN MUTANT FROM DESULFOVIBRIO VULGARIS | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Walsh, M.A, Mccarthy, A, O'Farrell, P.A, Voordouw, G, Higgins, T, Mayhew, S.G. | | Deposit date: | 1997-11-18 | | Release date: | 1998-05-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modulation of the redox potentials of FMN in Desulfovibrio vulgaris flavodoxin: thermodynamic properties and crystal structures of glycine-61 mutants.
Biochemistry, 37, 1998
|
|
1SRV
 
 | | THERMUS THERMOPHILUS GROEL (HSP60 CLASS) FRAGMENT (APICAL DOMAIN) COMPRISING RESIDUES 192-336 | | Descriptor: | PROTEIN (GROEL (HSP60 CLASS)) | | Authors: | Walsh, M.A, Dementieva, I, Evans, G, Sanishvili, R, Joachimiak, A. | | Deposit date: | 1999-03-02 | | Release date: | 1999-03-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Taking MAD to the extreme: ultrafast protein structure determination.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1DW9
 
 | | Structure of cyanase reveals that a novel dimeric and decameric arrangement of subunits is required for formation of the enzyme active site | | Descriptor: | CHLORIDE ION, CYANATE LYASE, SULFATE ION | | Authors: | Walsh, M.A, Otwinowski, Z, Perrakis, A, Anderson, P.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 1999-12-03 | | Release date: | 2000-05-16 | | Last modified: | 2019-08-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of Cyanase Reveals that a Novel Dimeric and Decameric Arrangement of Subunits is Required for Formation of the Enzyme Active Site
Structure, 8, 2000
|
|
1DWK
 
 | | STRUCTURE OF CYANASE WITH THE DI-ANION OXALATE BOUND AT THE ENZYME ACTIVE SITE | | Descriptor: | CYANATE HYDRATASE, OXALATE ION, SULFATE ION | | Authors: | Walsh, M.A, Otwinowski, Z, Perrakis, A, Anderson, P.M, Joachimiak, A. | | Deposit date: | 1999-12-07 | | Release date: | 2000-05-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of Cyanase Reveals that a Novel Dimeric and Decameric Arrangement of Subunits is Required for Formation of the Enzyme Active Site.
Structure, 8, 2000
|
|
4MCW
 
 | | Crystal structure of a HD-GYP domain (a cyclic-di-GMP phosphodiesterase) containing a tri-nuclear metal centre | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, IMIDAZOLE, ... | | Authors: | Bellini, D, Walsh, M.A, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2013-08-21 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure of an HD-GYP domain cyclic-di-GMP phosphodiesterase reveals an enzyme with a novel trinuclear catalytic iron centre.
Mol.Microbiol., 91, 2014
|
|
1HJO
 
 | | ATPase domain of human heat shock 70kDa protein 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Walsh, M.A, Freeman, B.C, Morimoto, R.I, Joachimiak, A. | | Deposit date: | 1998-10-13 | | Release date: | 1998-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a new crystal form of human Hsp70 ATPase domain.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
9FGO
 
 | | Crystal structure of Enterovirus 71 2A protease mutant C110A containing VP1-2A junction in the active site | | Descriptor: | CHLORIDE ION, Polyprotein, ZINC ION | | Authors: | Ni, X, Koekemoer, L, Williams, E.P, Wang, S, Wright, N.D, Godoy, A.S, Aschenbrenner, J.C, Balcomb, B.H, Lithgo, R.M, Marples, P.G, Fairhead, M, Thompson, W, Kirkegaard, K, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-05-24 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Crystal structure of Enterovirus 71 2A protease mutant C110A containing VP1-2A junction in the active site
To Be Published
|
|
6TR3
 
 | | Ruminococcus gnavus GH29 fucosidase E1_10125 in complex with fucose | | Descriptor: | CALCIUM ION, F5/8 type C domain-containing protein, MAGNESIUM ION, ... | | Authors: | Owen, C.D, Wu, H, Crost, E, Colvile, A, Juge, N, Walsh, M.A. | | Deposit date: | 2019-12-17 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Fucosidases from the human gut symbiont Ruminococcus gnavus.
Cell.Mol.Life Sci., 78, 2021
|
|
6TR4
 
 | | Ruminococcus gnavus GH29 fucosidase E1_10125 D221A mutant in complex with fucose | | Descriptor: | CALCIUM ION, CHLORIDE ION, F5/8 type C domain-containing protein, ... | | Authors: | Owen, C.D, Wu, H, Crost, E, Colvile, A, Juge, N, Walsh, M.A. | | Deposit date: | 2019-12-17 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Fucosidases from the human gut symbiont Ruminococcus gnavus.
Cell.Mol.Life Sci., 78, 2021
|
|
6AC6
 
 | | Ab initio crystal structure of Selenomethionine labelled Mycobacterium smegmatis Mfd | | Descriptor: | Mycobacterium smegmatis Mfd, SULFATE ION | | Authors: | Putta, S, Fox, G.C, Walsh, M.A, Rao, D.N, Nagaraja, V, Natesh, R. | | Deposit date: | 2018-07-25 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.989 Å) | | Cite: | Structural basis for nucleotide-mediated remodelling mechanism of Mycobacterium Mfd
To Be Published
|
|
6AC8
 
 | | Crystal structure of Mycobacterium smegmatis Mfd at 2.75 A resolution | | Descriptor: | Mycobacterium smegmatis Mfd, SULFATE ION | | Authors: | Putta, S, Fox, G.C, Walsh, M.A, Rao, D.N, Nagaraja, V, Natesh, R. | | Deposit date: | 2018-07-25 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for nucleotide-mediated remodelling mechanism of Mycobacterium Mfd
To Be Published
|
|
6ACX
 
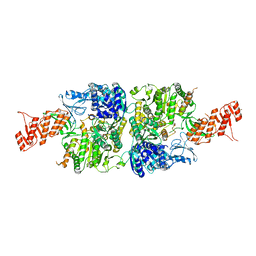 | | Crystal structure of Mycobacterium smegmatis Mfd in complex with ADP + Pi at 3.5 A resolution. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Mycobacterium smegmatis Mfd, PHOSPHATE ION, ... | | Authors: | Putta, S, Fox, G.C, Walsh, M.A, Rao, D.N, Nagaraja, V, Natesh, R. | | Deposit date: | 2018-07-27 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for nucleotide-mediated remodelling mechanism of Mycobacterium Mfd
To Be Published
|
|
6ACA
 
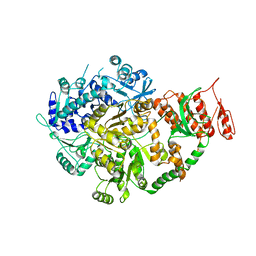 | | Crystal structure of Mycobacterium tuberculosis Mfd at 3.6 A resolution | | Descriptor: | Mycobacterium tuberculosis Mfd | | Authors: | Putta, S, Fox, G.C, Walsh, M.A, Rao, D.N, Nagaraja, V, Natesh, R. | | Deposit date: | 2018-07-26 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural basis for nucleotide-mediated remodelling mechanism of Mycobacterium Mfd
To Be Published
|
|
7Z4S
 
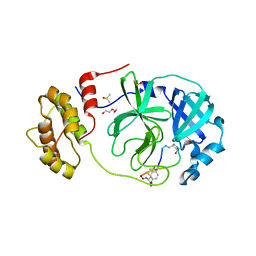 | | Crystal structure of SARS-CoV-2 Mpro in complex with cyclic peptide GM4 including unnatural amino acids. | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Owen, C.D, Miura, T, Malla, T, Lukacik, L, Strain-Damerell, C.M, Tumber, A, Brewitz, L, McDonough, M.A, Salah, E, Terasaka, N, Katoh, T, Kawamura, A, Schofield, C.J, Suga, H, Walsh, M.A. | | Deposit date: | 2022-03-04 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In vitro selection of macrocyclic peptide inhibitors containing cyclic gamma 2,4 -amino acids targeting the SARS-CoV-2 main protease.
Nat.Chem., 15, 2023
|
|
8UM3
 
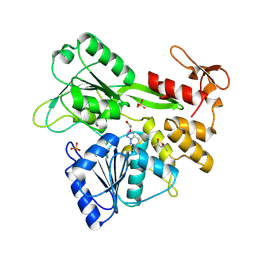 | | PanDDA analysis -- Crystal Structure of Zika virus NS3 Helicase in complex with Z203039992 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 6-chlorotetrazolo[1,5-b]pyridazine, ... | | Authors: | Godoy, A.S, Noske, G.D, Fairhead, M, Lithgo, R.M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Mesquita, N.C.M.R, Oliva, G, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-10-17 | | Release date: | 2023-11-01 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.925 Å) | | Cite: | PanDDA analysis -- Crystal Structure of Zika virus NS3 Helicase in complex with Z203039992
To Be Published
|
|
8V7R
 
 | | PanDDA analysis -- Crystal Structure of Zika virus NS3 Helicase in complex with Z56772132 | | Descriptor: | (5R)-5-[2-(4-methoxyphenyl)ethyl]-5-methylimidazolidine-2,4-dione, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Godoy, A.S, Noske, G.D, Fairhead, M, Lithgo, R.M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Mesquita, N.C.M.R, Oliva, G, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-12-04 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | PanDDA analysis -- Crystal Structure of Zika virus NS3 Helicase in complex with Z56772132
To Be Published
|
|
8V7U
 
 | | PanDDA analysis -- Crystal Structure of Zika virus NS3 Helicase in complex with Z729726784 | | Descriptor: | 1,2-ETHANEDIOL, 2-cyclopentyl-N-(3-methyl-1,2,4-oxadiazol-5-yl)acetamide, DIMETHYL SULFOXIDE, ... | | Authors: | Godoy, A.S, Noske, G.D, Fairhead, M, Lithgo, R.M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Mesquita, N.C.M.R, Oliva, G, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-12-04 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | PanDDA analysis -- Crystal Structure of Zika virus NS3 Helicase in complex with Z729726784
To Be Published
|
|
7AEG
 
 | | SARS-CoV-2 main protease in a covalent complex with SDZ 224015 derivative, compound 5 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[(benzyloxy)carbonyl]-L-valyl-N-[(1S)-1-(carboxymethyl)-3-fluoro-2-oxopropyl]-L-alaninamide | | Authors: | Owen, C.D, Redhead, M.A, Lukacik, P, Strain-Damerell, C, Fearon, D, Brewitz, L, Collette, A, Robinson, C, Collins, P, Radoux, C, Navratilova, I, Douangamath, A, von Delft, F, Malla, T.R, Nugen, T, Hull, H, Tumber, A, Schofield, C.J, Hallet, D, Stuart, D.I, Hopkins, A.L, Walsh, M.A. | | Deposit date: | 2020-09-17 | | Release date: | 2021-07-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Bispecific repurposed medicines targeting the viral and immunological arms of COVID-19.
Sci Rep, 11, 2021
|
|
7AEH
 
 | | SARS-CoV-2 main protease in a covalent complex with a pyridine derivative of ABT-957, compound 1 | | Descriptor: | (2~{R})-5-oxidanylidene-~{N}-[(2~{R},3~{S})-3-oxidanyl-4-oxidanylidene-1-phenyl-4-(pyridin-2-ylmethylamino)butan-2-yl]-1-(phenylmethyl)pyrrolidine-2-carboxamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Owen, C.D, Redhead, M.A, Lukacik, P, Strain-Damerell, C, Fearon, D, Brewitz, L, Collette, A, Robinson, C, Collins, P, Radoux, C, Navratilova, I, Douangamath, A, von Delft, F, Malla, T.R, Nugen, T, Hull, H, Tumber, A, Schofield, C.J, Hallet, D, Stuart, D.I, Hopkins, A.L, Walsh, M.A. | | Deposit date: | 2020-09-17 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Bispecific repurposed medicines targeting the viral and immunological arms of COVID-19.
Sci Rep, 11, 2021
|
|
7Z59
 
 | | SARS-CoV-2 main protease (Mpro) covalently modified with a penicillin derivative | | Descriptor: | (3S)-4-[[2,4-bis(fluoranyl)phenyl]methoxy]-2-methyl-4-oxidanylidene-3-[[(Z)-3-oxidanylidene-2-(2-phenoxyethanoylamino)prop-1-enyl]amino]butane-2-sulfinic acid, 1,2-ETHANEDIOL, 3C-like proteinase nsp5 | | Authors: | Owen, C.D, Malla, T.R, Brewitz, L, Lukacik, P, Strain-Damerell, C, Mikolajek, H, Muntean, D.G, Aslam, H, Salah, E, Tumber, A, Schofield, C.J, Walsh, M.A. | | Deposit date: | 2022-03-08 | | Release date: | 2022-06-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Penicillin Derivatives Inhibit the SARS-CoV-2 Main Protease by Reaction with Its Nucleophilic Cysteine.
J.Med.Chem., 65, 2022
|
|
8B2T
 
 | | SARS-CoV-2 Main Protease (Mpro) in complex with nirmatrelvir alkyne | | Descriptor: | 3C-like proteinase nsp5, Nirmatrelvir (reacted form) | | Authors: | Owen, C.D, Crawshaw, A.D, Warren, A.J, Trincao, J, Zhao, Y, Brewitz, L, Malla, T.R, Salah, E, Petra, L, Strain-Damerell, C, Schofield, C.J, Walsh, M.A. | | Deposit date: | 2022-09-14 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | Alkyne Derivatives of SARS-CoV-2 Main Protease Inhibitors Including Nirmatrelvir Inhibit by Reacting Covalently with the Nucleophilic Cysteine.
J.Med.Chem., 66, 2023
|
|
6YB7
 
 | | SARS-CoV-2 main protease with unliganded active site (2019-nCoV, coronavirus disease 2019, COVID-19). | | Descriptor: | 3C-like proteinase, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE | | Authors: | Owen, C.D, Lukacik, P, Strain-Damerell, C.M, Douangamath, A, Powell, A.J, Fearon, D, Brandao-Neto, J, Crawshaw, A.D, Aragao, D, Williams, M, Flaig, R, Hall, D.R, McAuley, K.E, Mazzorana, M, Stuart, D.I, von Delft, F, Walsh, M.A. | | Deposit date: | 2020-03-16 | | Release date: | 2020-03-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | COVID-19 main protease with unliganded active site
To Be Published
|
|
