3CGD
 
 | |
3CGC
 
 | |
8QH6
 
 | | Crystal structure of IpgC in complex with a follow-up compound based on J20 | | Descriptor: | 3-azanyl-6-chloranyl-isoindol-1-one, CHLORIDE ION, Chaperone protein IpgC, ... | | Authors: | Wallbaum, J.E, Heine, A, Reuter, K. | | Deposit date: | 2023-09-06 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic Fragment Screening on the Shigella Type III Secretion System Chaperone IpgC.
Acs Omega, 8, 2023
|
|
3ICS
 
 | |
3ICR
 
 | | Crystal structure of oxidized Bacillus anthracis CoADR-RHD | | Descriptor: | COENZYME A, Coenzyme A-Disulfide Reductase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Wallen, J.R, Claiborne, A. | | Deposit date: | 2009-07-18 | | Release date: | 2009-11-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and catalytic properties of Bacillus anthracis CoADR-RHD: implications for flavin-linked sulfur trafficking.
Biochemistry, 48, 2009
|
|
3ICT
 
 | | Crystal structure of reduced Bacillus anthracis CoADR-RHD | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, COENZYME A, Coenzyme A-Disulfide Reductase, ... | | Authors: | Wallen, J.R, Claiborne, A. | | Deposit date: | 2009-07-18 | | Release date: | 2009-11-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and catalytic properties of Bacillus anthracis CoADR-RHD: implications for flavin-linked sulfur trafficking.
Biochemistry, 48, 2009
|
|
5IKN
 
 | |
2HAV
 
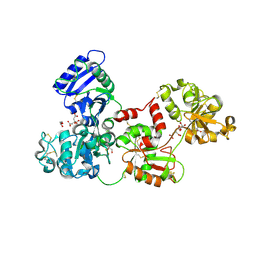 | | Apo-Human Serum Transferrin (Glycosylated) | | Descriptor: | CITRIC ACID, GLYCEROL, Serotransferrin | | Authors: | Wally, J, Everse, S.J. | | Deposit date: | 2006-06-13 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Crystal Structure of Iron-free Human Serum Transferrin Provides Insight into Inter-lobe Communication and Receptor Binding.
J.Biol.Chem., 281, 2006
|
|
2HAU
 
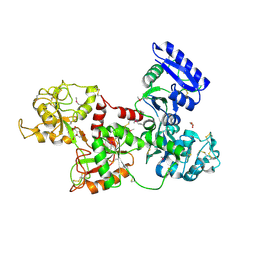 | | Apo-Human Serum Transferrin (Non-Glycosylated) | | Descriptor: | CITRIC ACID, GLYCEROL, Serotransferrin | | Authors: | Wally, J, Everse, S.J. | | Deposit date: | 2006-06-13 | | Release date: | 2006-06-27 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Crystal Structure of Iron-free Human Serum Transferrin Provides Insight into Inter-lobe Communication and Receptor Binding.
J.Biol.Chem., 281, 2006
|
|
3CGE
 
 | |
3CGB
 
 | |
3IYO
 
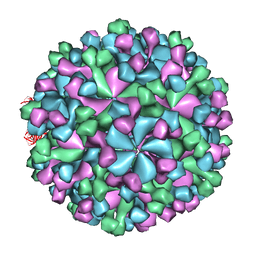 | | Cryo-EM model of virion-sized HEV virion-sized capsid | | Descriptor: | Capsid protein | | Authors: | Xing, L, Mayazaki, N, Li, T.C, Simons, M.N, Wall, J.S, Moore, M, Wang, C.Y, Takeda, N, Wakita, T, Miyamura, T, Cheng, R.H. | | Deposit date: | 2010-03-19 | | Release date: | 2010-08-18 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (10.5 Å) | | Cite: | Structural basis for the RNA-dependent assembly pathway of hepatitis E virion-sized particles
J.Biol.Chem., 2010
|
|
5KK3
 
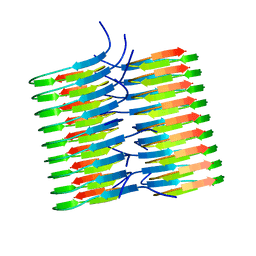 | | Atomic Resolution Structure of Monomorphic AB42 Amyloid Fibrils | | Descriptor: | Beta-amyloid protein 42 | | Authors: | Colvin, M.T, Silvers, R, Zhe Ni, Q, Can, T.V, Sergeyev, I, Rosay, M, Donovan, K.J, Michael, B, Wall, J, Linse, S, Griffin, R.G. | | Deposit date: | 2016-06-20 | | Release date: | 2016-07-13 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Atomic Resolution Structure of Monomorphic A beta 42 Amyloid Fibrils.
J.Am.Chem.Soc., 138, 2016
|
|
6EKA
 
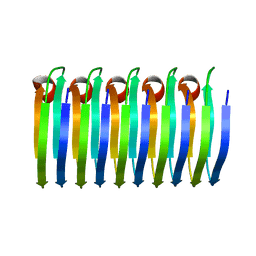 | | Solid-state MAS NMR structure of the HELLF prion amyloid fibrils | | Descriptor: | Podospora anserina S mat+ genomic DNA chromosome 3, supercontig 2 | | Authors: | Martinez, D, Daskalov, A, Andreas, L, Bardiaux, B, Coustou, V, Stanek, J, Berbon, M, Noubhani, M, Kauffmann, B, Wall, J.S, Pintacuda, G, Saupe, S.J, Habenstein, B, Loquet, A. | | Deposit date: | 2017-09-25 | | Release date: | 2018-10-10 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | Structural and molecular basis of cross-seeding barriers in amyloids
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
2A3P
 
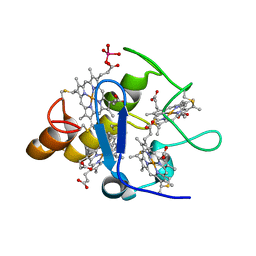 | | Structure of Desulfovibrio desulfuricans G20 tetraheme cytochrome with bound molybdate | | Descriptor: | COG3005: Nitrate/TMAO reductases, membrane-bound tetraheme cytochrome c subunit, HEME C, ... | | Authors: | Pattarkine, M.V, Lee, Y.-H, Tanner, J.J, Wall, J.D. | | Deposit date: | 2005-06-25 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Desulfovibrio desulfuricans G20 Tetraheme Cytochrome Structure at 1.5A and Cytochrome Interaction with Metal Complexes
J.Mol.Biol., 358, 2006
|
|
2A3M
 
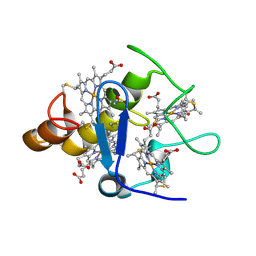 | | Structure of Desulfovibrio desulfuricans G20 tetraheme cytochrome (oxidized form) | | Descriptor: | COG3005: Nitrate/TMAO reductases, membrane-bound tetraheme cytochrome c subunit, HEME C | | Authors: | Pattarkine, M.V, Tanner, J.J, Bottoms, C.A, Lee, Y.H, Wall, J.D. | | Deposit date: | 2005-06-25 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Desulfovibrio desulfuricans G20 Tetraheme Cytochrome Structure at 1.5A and Cytochrome Interaction with Metal Complexes
J.Mol.Biol., 358, 2006
|
|
2NAO
 
 | | Atomic resolution structure of a disease-relevant Abeta(1-42) amyloid fibril | | Descriptor: | Beta-amyloid protein 42 | | Authors: | Waelti, M.A, Ravotti, F, Arai, H, Glabe, C, Wall, J, Bockmann, A, Guntert, P, Meier, B.H, Riek, R. | | Deposit date: | 2016-01-07 | | Release date: | 2016-07-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Atomic-resolution structure of a disease-relevant A beta (1-42) amyloid fibril.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2MVX
 
 | | Atomic-resolution 3D structure of amyloid-beta fibrils: the Osaka mutation | | Descriptor: | Amyloid beta A4 protein | | Authors: | Schuetz, A.K, Vagt, T, Huber, M, Ovchinnikova, O.Y, Cadalbert, R, Wall, J, Guentert, P, Bockmann, A, Glockshuber, R, Meier, B.H. | | Deposit date: | 2014-10-17 | | Release date: | 2014-11-26 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Atomic-Resolution Three-Dimensional Structure of Amyloid beta Fibrils Bearing the Osaka Mutation.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
3LRI
 
 | | Solution structure and backbone dynamics of long-[Arg(3)]insulin-like growth factor-I | | Descriptor: | PROTEIN (INSULIN-LIKE GROWTH FACTOR I) | | Authors: | Laajoki, L.G, Francis, G.L, Wallace, J.C, Carver, J.A, Keniry, M.A. | | Deposit date: | 1999-04-13 | | Release date: | 2000-05-23 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of long-[Arg(3)]insulin-like growth factor-I
J.Biol.Chem., 275, 2000
|
|
5W0O
 
 | |
5W0N
 
 | | Structure of human TUT7 catalytic module (CM) in complex with UMPNPP and U2 RNA | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, IODIDE ION, MAGNESIUM ION, ... | | Authors: | Faehnle, C.R, Walleshauser, J, Joshua-Tor, L. | | Deposit date: | 2017-05-31 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Multi-domain utilization by TUT4 and TUT7 in control of let-7 biogenesis.
Nat. Struct. Mol. Biol., 24, 2017
|
|
4PMW
 
 | |
6P7E
 
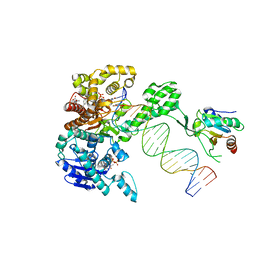 | | Structure of T7 DNA Polymerase Bound to a Primer/Template DNA and a Peptide that Mimics the C-terminal Tail of the Primase-Helicase | | Descriptor: | ASP-THR-ASP-PHE peptide, DNA (25-MER), DNA (5'-D(P*GP*GP*CP*AP*GP*GP*TP*GP*GP*TP*CP*TP*TP*GP*CP*CP*GP*GP*TP*GP*A)-3'), ... | | Authors: | Foster, B.M, Rosenberg, D, Salvo, H, Stephens, K.L, Bintz, B.J, Hammel, M, Ellenberger, T, Gainey, M.D, Wallen, J.R. | | Deposit date: | 2019-06-05 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Combined Solution and Crystal Methods Reveal the Electrostatic Tethers That Provide a Flexible Platform for Replication Activities in the Bacteriophage T7 Replisome.
Biochemistry, 58, 2019
|
|
7TZ1
 
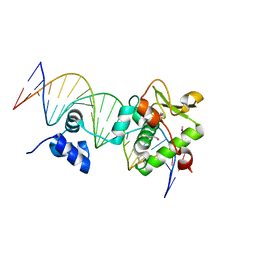 | | Crystal Structure of a Mycobacteriophage Cluster A2 Immunity Repressor:DNA Complex | | Descriptor: | DNA (5'-D(P*CP*CP*CP*GP*CP*TP*TP*GP*AP*CP*AP*GP*CP*CP*AP*CP*CP*GP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*CP*GP*GP*TP*GP*GP*CP*TP*GP*TP*CP*AP*AP*GP*CP*GP*GP*G)-3'), Immunity repressor | | Authors: | McGinnis, R.J, Brambley, C.A, Stamey, B, Green, W.C, Gragg, K.N, Cafferty, E.R, Terwilliger, T.C, Hammel, M, Hollis, T.J, Miller, J.M, Gainey, M.D, Wallen, J.R. | | Deposit date: | 2022-02-15 | | Release date: | 2022-07-20 | | Last modified: | 2022-07-27 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | A monomeric mycobacteriophage immunity repressor utilizes two domains to recognize an asymmetric DNA sequence.
Nat Commun, 13, 2022
|
|
7R6R
 
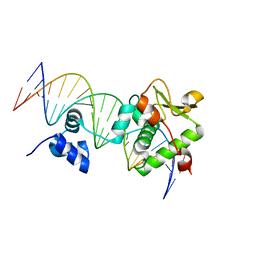 | | Crystal Structure of a Mycobacteriophage Cluster A2 Immunity Repressor:DNA Complex | | Descriptor: | DNA (5'-D(P*CP*CP*CP*GP*CP*TP*TP*GP*AP*CP*AP*GP*CP*CP*AP*CP*CP*GP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*CP*GP*GP*TP*GP*GP*CP*TP*GP*TP*CP*AP*AP*GP*CP*GP*GP*G)-3'), Immunity repressor | | Authors: | McGinnis, R.J, Brambley, C.A, Stamey, B, Green, W.C, Gragg, K.N, Cafferty, E.R, Terwilliger, T.C, Hammel, M, Hollis, T.J, Miller, J.M, Gainey, M.D, Wallen, J.R. | | Deposit date: | 2021-06-23 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | A monomeric mycobacteriophage immunity repressor utilizes two domains to recognize an asymmetric DNA sequence.
Nat Commun, 13, 2022
|
|
