6ZNQ
 
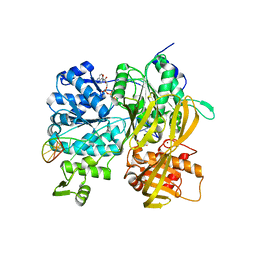 | | Crystal Structure of DUF1998 helicase MrfA bound to DNA and AMPPNP | | Descriptor: | CITRIC ACID, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Uncharacterized ATP-dependent helicase YprA, ... | | Authors: | Roske, J.J, Liu, S, Loll, B, Neu, U, Wahl, M.C. | | Deposit date: | 2020-07-06 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | A skipping rope translocation mechanism in a widespread family of DNA repair helicases.
Nucleic Acids Res., 49, 2021
|
|
6ZNS
 
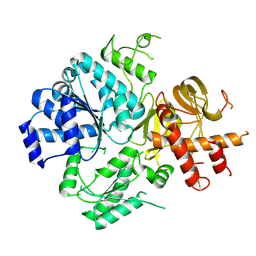 | | Crystal Structure of DUF1998 helicase MrfA | | Descriptor: | Uncharacterized ATP-dependent helicase YprA, ZINC ION | | Authors: | Roske, J.J, Liu, S, Loll, B, Neu, U, Wahl, M.C. | | Deposit date: | 2020-07-06 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | A skipping rope translocation mechanism in a widespread family of DNA repair helicases.
Nucleic Acids Res., 49, 2021
|
|
6ZWX
 
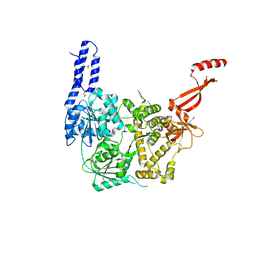 | | Crystal structure of E. coli RNA helicase HrpA | | Descriptor: | ATP-dependent RNA helicase HrpA | | Authors: | Grass, L.M, Wollenhaupt, J, Barthel, T, Loll, B, Wahl, M.C. | | Deposit date: | 2020-07-29 | | Release date: | 2021-06-30 | | Last modified: | 2021-08-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Large-scale ratcheting in a bacterial DEAH/RHA-type RNA helicase that modulates antibiotics susceptibility.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6ZWW
 
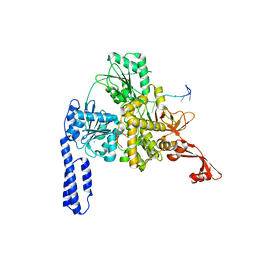 | | Crystal structure of E. coli RNA helicase HrpA in complex with RNA | | Descriptor: | ATP-dependent RNA helicase HrpA, CALCIUM ION, ssRNA | | Authors: | Grass, L.M, Wollenhaupt, J, Barthel, T, Loll, B, Wahl, M.C. | | Deposit date: | 2020-07-29 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Large-scale ratcheting in a bacterial DEAH/RHA-type RNA helicase that modulates antibiotics susceptibility.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7AKP
 
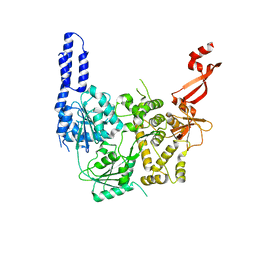 | | Crystal structure of E. coli RNA helicase HrpA-D305A | | Descriptor: | ATP-dependent RNA helicase HrpA | | Authors: | Grass, L.M, Wollenhaupt, J, Barthel, T, Loll, B, Wahl, M.C. | | Deposit date: | 2020-10-01 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Large-scale ratcheting in a bacterial DEAH/RHA-type RNA helicase that modulates antibiotics susceptibility.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7B3A
 
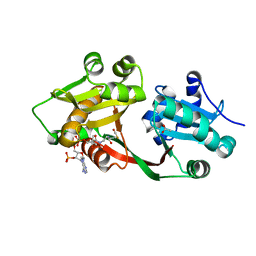 | | Crystal structure of PamZ | | Descriptor: | ACETATE ION, ACETYL COENZYME *A, CHLORIDE ION, ... | | Authors: | Loll, B, Dang, T, Mainz, A, Suessmuth, R, Wahl, M.C. | | Deposit date: | 2020-11-30 | | Release date: | 2021-11-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Molecular basis of antibiotic self-resistance in a bee larvae pathogen.
Nat Commun, 13, 2022
|
|
4F92
 
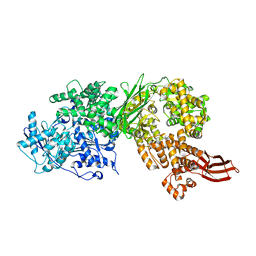 | | Brr2 Helicase Region S1087L | | Descriptor: | SULFANILAMIDE, U5 small nuclear ribonucleoprotein 200 kDa helicase | | Authors: | Santos, K.F, Jovin, S.M, Weber, G, Pena, V, Luehrmann, R, Wahl, M.C. | | Deposit date: | 2012-05-18 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.662 Å) | | Cite: | Structural basis for functional cooperation between tandem helicase cassettes in Brr2-mediated remodeling of the spliceosome.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4F91
 
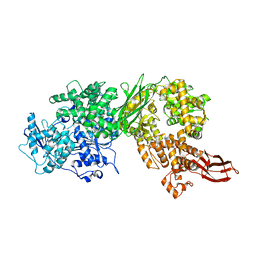 | | Brr2 Helicase Region | | Descriptor: | U5 small nuclear ribonucleoprotein 200 kDa helicase | | Authors: | Santos, K.F, Jovin, S.M, Weber, G, Pena, V, Luehrmann, R, Wahl, M.C. | | Deposit date: | 2012-05-18 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.697 Å) | | Cite: | Structural basis for functional cooperation between tandem helicase cassettes in Brr2-mediated remodeling of the spliceosome.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4F93
 
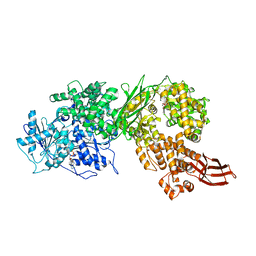 | | Brr2 Helicase Region S1087L, Mg-ATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Santos, K.F, Jovin, S.M, Weber, G, Pena, V, Luehrmann, R, Wahl, M.C. | | Deposit date: | 2012-05-18 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Structural basis for functional cooperation between tandem helicase cassettes in Brr2-mediated remodeling of the spliceosome.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
438D
 
 | |
3HD7
 
 | | HELICAL EXTENSION OF THE NEURONAL SNARE COMPLEX INTO THE MEMBRANE, spacegroup C 1 2 1 | | Descriptor: | SULFATE ION, Synaptosomal-associated protein 25, Syntaxin-1A, ... | | Authors: | Stein, A, Weber, G, Wahl, M.C, Jahn, R. | | Deposit date: | 2009-05-07 | | Release date: | 2009-07-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Helical extension of the neuronal SNARE complex into the membrane
Nature, 460, 2009
|
|
3IM2
 
 | |
3IMQ
 
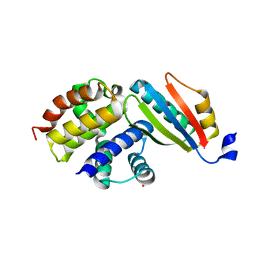 | | Crystal structure of the NusB101-S10(delta loop) complex | | Descriptor: | 30S ribosomal protein S10, N utilization substance protein B, POTASSIUM ION | | Authors: | Luo, X, Wahl, M.C. | | Deposit date: | 2009-08-11 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fine tuning of the E. coli NusB:NusE complex affinity to BoxA RNA is required for processive antitermination.
Nucleic Acids Res., 38, 2010
|
|
3IM1
 
 | |
3IPD
 
 | | Helical extension of the neuronal SNARE complex into the membrane, spacegroup I 21 21 21 | | Descriptor: | Synaptosomal-associated protein 25, Syntaxin-1A, Vesicle-associated membrane protein 2 | | Authors: | Stein, A, Weber, G, Wahl, M.C, Jahn, R. | | Deposit date: | 2009-08-17 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (4.8 Å) | | Cite: | Helical extension of the neuronal SNARE complex into the membrane
Nature, 460, 2009
|
|
3LSA
 
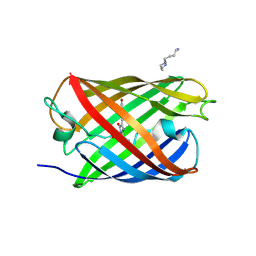 | | Padron0.9-OFF (non-fluorescent state) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, Padron0.9, ... | | Authors: | Brakemann, T, Weber, G, Trowitzsch, S, Wahl, M.C, Jakobs, S. | | Deposit date: | 2010-02-12 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Molecular basis of the light-driven switching of the photochromic fluorescent protein Padron.
J.Biol.Chem., 285, 2010
|
|
3LS3
 
 | | Padron0.9-ON (fluorescent state) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, Padron0.9, ... | | Authors: | Brakemann, T, Weber, G, Trowitzsch, S, Wahl, M.C, Jakobs, S. | | Deposit date: | 2010-02-12 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Molecular basis of the light-driven switching of the photochromic fluorescent protein Padron.
J.Biol.Chem., 285, 2010
|
|
4PSO
 
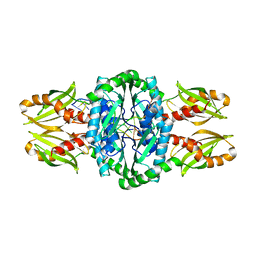 | | Crystal structure of apeThermo-DBP-RP2 bound to ssDNA dT10 | | Descriptor: | PHOSPHATE ION, polydeoxyribonucleotide, ssDNA binding protein | | Authors: | Gahlei, H, von Moeller, H, Eppers, D, Loll, B, Wahl, M.C. | | Deposit date: | 2014-03-07 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Entrapment of DNA in an intersubunit tunnel system of a single-stranded DNA-binding protein.
Nucleic Acids Res., 42, 2014
|
|
4PSM
 
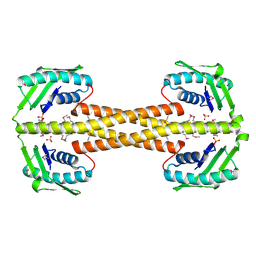 | | Crystal structure of pfuThermo-DBP-RP1 (crystal form II) | | Descriptor: | SULFATE ION, ssDNA binding protein | | Authors: | Gahlei, H, von Moeller, H, Eppers, D, Loll, B, Wahl, M.C. | | Deposit date: | 2014-03-07 | | Release date: | 2014-04-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Entrapment of DNA in an intersubunit tunnel system of a single-stranded DNA-binding protein.
Nucleic Acids Res., 42, 2014
|
|
4PSL
 
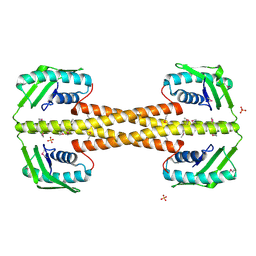 | | Crystal structure of pfuThermo-DBP-RP1 (crystal form I) | | Descriptor: | SULFATE ION, ssDNA binding protein | | Authors: | Gahlei, H, von Moeller, H, Eppers, D, Sohmen, D, Wilson, D.N, Loll, B, Wahl, M.C. | | Deposit date: | 2014-03-07 | | Release date: | 2014-04-30 | | Last modified: | 2014-06-25 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Entrapment of DNA in an intersubunit tunnel system of a single-stranded DNA-binding protein.
Nucleic Acids Res., 42, 2014
|
|
4PSN
 
 | | Crystal structure of apeThermo-DBP-RP2 | | Descriptor: | GLYCEROL, IMIDAZOLE, ssDNA binding protein | | Authors: | Gahlei, H, von Moeller, H, Eppers, D, Loll, B, Wahl, M.C. | | Deposit date: | 2014-03-07 | | Release date: | 2014-04-30 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Entrapment of DNA in an intersubunit tunnel system of a single-stranded DNA-binding protein.
Nucleic Acids Res., 42, 2014
|
|
4R3E
 
 | |
4R3F
 
 | | Structure of the spliceosomal peptidyl-prolyl cis-trans isomerase Cwc27 from Chaetomium thermophilum | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ACETATE ION, ... | | Authors: | Ulrich, A, Wahl, M.C. | | Deposit date: | 2014-08-15 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure and evolution of the spliceosomal peptidyl-prolyl cis-trans isomerase Cwc27.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3D3C
 
 | |
3D3B
 
 | |
