3NN1
 
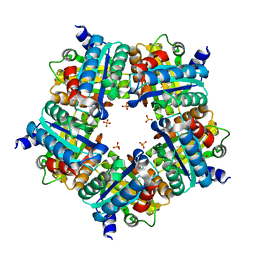 | | Structure of chlorite dismutase from Candidatus Nitrospira defluvii in complex with imidazole | | Descriptor: | 1,2-ETHANEDIOL, Chlorite dismutase, IMIDAZOLE, ... | | Authors: | Kostan, J, Sjoeblom, B, Maixner, F, Mlynek, G, Furtmueller, P.G, Obinger, C, Wagner, M, Daims, H, Djinovic-Carugo, K. | | Deposit date: | 2010-06-23 | | Release date: | 2010-07-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and functional characterisation of the chlorite dismutase from the nitrite-oxidizing bacterium "Candidatus Nitrospira defluvii": Identification of a catalytically important amino acid residue
J.Struct.Biol., 172, 2010
|
|
3NN3
 
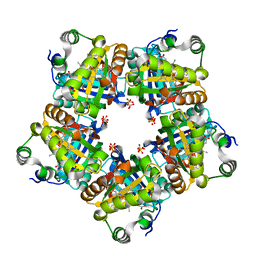 | | Structure of chlorite dismutase from Candidatus Nitrospira defluvii R173A mutant | | Descriptor: | Chlorite dismutase, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Kostan, J, Sjoeblom, B, Maixner, F, Mlynek, G, Furtmueller, P.G, Obinger, C, Wagner, M, Daims, H, Djinovic-Carugo, K. | | Deposit date: | 2010-06-23 | | Release date: | 2010-07-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional characterisation of the chlorite dismutase from the nitrite-oxidizing bacterium "Candidatus Nitrospira defluvii": Identification of a catalytically important amino acid residue
J.Struct.Biol., 172, 2010
|
|
3NN4
 
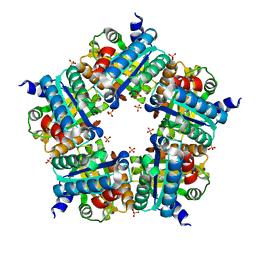 | | Structure of chlorite dismutase from Candidatus Nitrospira defluvii R173K mutant | | Descriptor: | Chlorite dismutase, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Kostan, J, Sjoeblom, B, Maixner, F, Mlynek, G, Furtmueller, P.G, Obinger, C, Wagner, M, Daims, H, Djinovic-Carugo, K. | | Deposit date: | 2010-06-23 | | Release date: | 2010-07-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and functional characterisation of the chlorite dismutase from the nitrite-oxidizing bacterium "Candidatus Nitrospira defluvii": Identification of a catalytically important amino acid residue
J.Struct.Biol., 172, 2010
|
|
4A7I
 
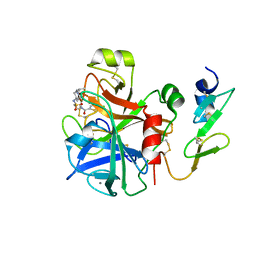 | | Factor Xa in complex with a potent 2-amino-ethane sulfonamide inhibitor | | Descriptor: | 5-CHLORO-THIOPHENE-2-CARBOXYLIC ACID [2-(1--ISOPROPYL-PIPERIDIN-4-YLSULFAMOYL)-ETHYL]-AMIDE, ACTIVATED FACTOR XA HEAVY CHAIN XA, CALCIUM ION, ... | | Authors: | Nazare, M, Matter, H, Will, D.W, Wagner, M, Urmann, M, Czech, J, Schreuder, H, Bauer, A, Ritter, K, Wehner, V. | | Deposit date: | 2011-11-14 | | Release date: | 2012-02-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Fragment Deconstruction of Small, Potent Factor Xa Inhibitors: Exploring the Superadditivity Energetics of Fragment Linking in Protein-Ligand Complexes.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
2GB0
 
 | | Monomeric sarcosine oxidase: structure of a covalently flavinylated amine oxidizing enzyme | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Monomeric sarcosine oxidase, ... | | Authors: | Trickey, P, Wagner, M.A, Jorns, M.S, Mathews, F.S. | | Deposit date: | 2006-03-09 | | Release date: | 2006-03-21 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Monomeric Sarcosine Oxidase: Structure of a Covalently Flavinylated Amine Oxidizing Enzyme
Structure, 7, 1999
|
|
