1RZ2
 
 | | 1.6A crystal structure of the protein BA4783/Q81L49 (similar to sortase B) from Bacillus anthracis. | | Descriptor: | conserved hypothetical protein BA4783 | | Authors: | Wu, R, Zhang, R, Gornicki, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-12-23 | | Release date: | 2004-07-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of sortase B from Staphylococcus aureus and Bacillus anthracis reveal catalytic amino acid triad in the active site.
Structure, 12, 2004
|
|
2QVZ
 
 | | 4-Chlorobenzoyl-CoA Ligase/Synthetase, I303A mutation, bound to 3-Chlorobenzoate | | Descriptor: | 3-chlorobenzoate, 4-Chlorobenzoate CoA Ligase/Synthetase | | Authors: | Wu, R, Reger, A.S, Cao, J, Gulick, A.M, Dunaway-Mariano, D. | | Deposit date: | 2007-08-09 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Rational redesign of the 4-chlorobenzoate binding site of 4-chlorobenzoate: coenzyme a ligase for expanded substrate range.
Biochemistry, 46, 2007
|
|
1RLJ
 
 | | Structural Genomics, a Flavoprotein NrdI from Bacillus subtilis | | Descriptor: | FLAVIN MONONUCLEOTIDE, IODIDE ION, NrdI protein | | Authors: | Wu, R, Zhang, R, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-11-25 | | Release date: | 2004-07-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 1.5A crystal structure of a thioredoxin-like protein NrdI from Bacillus subtilis
To be Published
|
|
4ZGJ
 
 | | Double Mutant H80A/H81A of Fe-Type Nitrile Hydratase from Comamonas testosteroni Ni1 | | Descriptor: | FE (III) ION, Nitrile hydratase alpha subunit, Nitrile hydratase beta subunit | | Authors: | Wu, R, Martinez, S, Holz, R, Liu, D. | | Deposit date: | 2015-04-23 | | Release date: | 2015-07-01 | | Last modified: | 2015-07-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analyzing the catalytic role of active site residues in the Fe-type nitrile hydratase from Comamonas testosteroni Ni1.
J.Biol.Inorg.Chem., 20, 2015
|
|
4WKV
 
 | | n-Alkylboronic Acid Inhibitors Reveal Determinants of Ligand Specificity in the Quorum-Quenching and Siderophore Biosynthetic Enzyme PvdQ | | Descriptor: | Acyl-homoserine lactone acylase PvdQ, GLYCEROL, trihydroxy(octyl)borate(1-) | | Authors: | Wu, R, Clevenger, K.D, Fast, W, Liu, D. | | Deposit date: | 2014-10-03 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1434 Å) | | Cite: | n-Alkylboronic Acid Inhibitors Reveal Determinants of Ligand Specificity in the Quorum-Quenching and Siderophore Biosynthetic Enzyme PvdQ.
Biochemistry, 53, 2014
|
|
4ZGD
 
 | | Mutant R157A of Fe-Type Nitrile Hydratase from Comamonas testosteroni Ni1 | | Descriptor: | FE (III) ION, Nitrile hydratase alpha subunit, Nitrile hydratase beta subunit | | Authors: | Wu, R, Martinez, S, Holz, R, Liu, D. | | Deposit date: | 2015-04-22 | | Release date: | 2015-07-01 | | Last modified: | 2015-07-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Analyzing the catalytic role of active site residues in the Fe-type nitrile hydratase from Comamonas testosteroni Ni1.
J.Biol.Inorg.Chem., 20, 2015
|
|
4ZSW
 
 | | Pig Brain GABA-AT inactivated by (E)-(1S,3S)-3-Amino-4-fluoromethylenyl-1-cyclopentanoic acid | | Descriptor: | (1S)-4-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]cyclopent-3-ene-1,3-dicarboxylic acid, 4-aminobutyrate aminotransferase, mitochondrial, ... | | Authors: | Wu, R, Lee, H, Le, H.V, Doud, E, Sanishvili, R, Compton, P, Kelleher, N.L, Silverman, R.B, Liu, D. | | Deposit date: | 2015-05-14 | | Release date: | 2015-07-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of Inactivation of GABA Aminotransferase by (E)- and (Z)-(1S,3S)-3-Amino-4-fluoromethylenyl-1-cyclopentanoic Acid.
Acs Chem.Biol., 10, 2015
|
|
4WKU
 
 | | n-Alkylboronic Acid Inhibitors Reveal Determinants of Ligand Specificity in the Quorum-Quenching and Siderophore Biosynthetic Enzyme PvdQ | | Descriptor: | Acyl-homoserine lactone acylase PvdQ, GLYCEROL, hexyl(trihydroxy)borate(1-) | | Authors: | Wu, R, Clevenger, K.D, Fast, W, Liu, D. | | Deposit date: | 2014-10-03 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | n-Alkylboronic Acid Inhibitors Reveal Determinants of Ligand Specificity in the Quorum-Quenching and Siderophore Biosynthetic Enzyme PvdQ.
Biochemistry, 53, 2014
|
|
4WKT
 
 | | n-Alkylboronic Acid Inhibitors Reveal Determinants of Ligand Specificity in the Quorum-Quenching and Siderophore Biosynthetic Enzyme PvdQ | | Descriptor: | 1-BUTANE BORONIC ACID, Acyl-homoserine lactone acylase PvdQ, GLYCEROL | | Authors: | Wu, R, Clevenger, K.D, Fast, W, Liu, D. | | Deposit date: | 2014-10-03 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.782 Å) | | Cite: | n-Alkylboronic Acid Inhibitors Reveal Determinants of Ligand Specificity in the Quorum-Quenching and Siderophore Biosynthetic Enzyme PvdQ.
Biochemistry, 53, 2014
|
|
4ZGE
 
 | | Double Mutant H80W/H81W of Fe-Type Nitrile Hydratase from Comamonas testosteroni Ni1 | | Descriptor: | FE (III) ION, Nitrile hydratase alpha subunit, Nitrile hydratase beta subunit | | Authors: | Wu, R, Martinez, S, Holz, R, Liu, D. | | Deposit date: | 2015-04-22 | | Release date: | 2015-07-01 | | Last modified: | 2015-07-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Analyzing the catalytic role of active site residues in the Fe-type nitrile hydratase from Comamonas testosteroni Ni1.
J.Biol.Inorg.Chem., 20, 2015
|
|
3K32
 
 | |
4ZSY
 
 | | Pig Brain GABA-AT inactivated by (Z)-(1S,3S)-3-Amino-4-fluoromethylenyl-1-cyclopentanoic acid. | | Descriptor: | (1S)-4-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]cyclopent-3-ene-1,3-dicarboxylic acid, 4-aminobutyrate aminotransferase, mitochondrial, ... | | Authors: | Wu, R, Lee, H, Le, H.V, Doud, E, Sanishvili, R, Compton, P, Kelleher, N.L, Silverman, R.B, Liu, D. | | Deposit date: | 2015-05-14 | | Release date: | 2015-07-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of Inactivation of GABA Aminotransferase by (E)- and (Z)-(1S,3S)-3-Amino-4-fluoromethylenyl-1-cyclopentanoic Acid.
Acs Chem.Biol., 10, 2015
|
|
8SQB
 
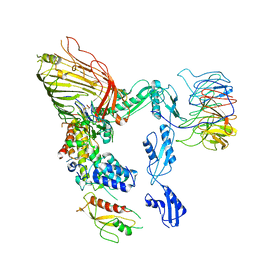 | | The cryo-EM structure of the EcBAM/EspP(beta7-12) complex | | Descriptor: | Maltodextrin-binding protein,EspP(b7-12), Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Wu, R, Noinaj, N. | | Deposit date: | 2023-05-04 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | BAM orchestrates OMP biogenesis using a beta-templating mechanism
To Be Published
|
|
8SQA
 
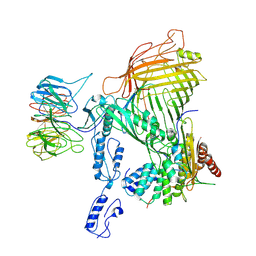 | | The cryo-EM structure of the EcBAM/EspP(beta8-12) complex | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Autotransporter protein EspP translocator, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Wu, R, Noinaj, N. | | Deposit date: | 2023-05-04 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | BAM orchestrates OMP biogenesis using a beta-templating mechanism
To Be Published
|
|
8SPR
 
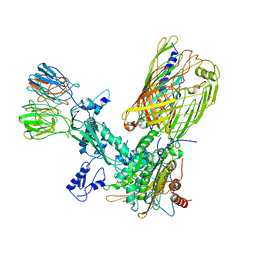 | | The cryo-EM structure of the EcBAM/EspP(beta1-12) complex | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Wu, R, Noinaj, N. | | Deposit date: | 2023-05-03 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | BAM orchestrates OMP biogenesis using a beta-templating mechanism
To Be Published
|
|
4MGR
 
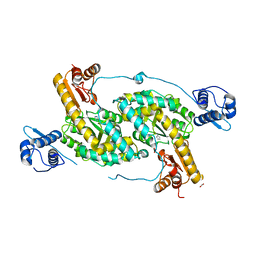 | | The crystal structure of Bacillus subtilis GabR, an autorepressor and PLP- and GABA-dependent transcriptional activator of gabT | | Descriptor: | ACETATE ION, HTH-type transcriptional regulatory protein GabR, IMIDAZOLE, ... | | Authors: | Wu, R, Edayathumangalam, R, Garcia, R, Wang, Y, Wang, W, Kreinbring, C.A, Bach, A, Liao, J, Stone, T, Terwilliger, T, Hoang, Q.Q, Belitsky, B.R, Petsko, G.A, Ringe, D, Liu, D. | | Deposit date: | 2013-08-28 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of Bacillus subtilis GabR, an autorepressor and transcriptional activator of gabT.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4EW7
 
 | | The crystal structure of conjugative transfer PAS_like domain from Salmonella enterica subsp. enterica serovar Typhimurium | | Descriptor: | ACETIC ACID, CHLORIDE ION, Conjugative transfer: regulation, ... | | Authors: | Wu, R, Jedrzejczak, R.P, Brown, R.N, Cort, J.R, Heffron, F, Nakayasu, E.S, Adkins, J.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Program for the Characterization of Secreted Effector Proteins (PCSEP) | | Deposit date: | 2012-04-26 | | Release date: | 2012-09-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The crystal structure of conjugative transfer PAS_like domain from Salmonella enterica subsp. enterica serovar Typhimurium
To be Published
|
|
4JGQ
 
 | | The crystal structure of sporulation kinase D mutant sensor domain, r131a, from Bacillus subtilis subsp in co-crystallization with pyruvate | | Descriptor: | ACETIC ACID, Sporulation kinase D | | Authors: | Wu, R, Schiffer, M, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-03-01 | | Release date: | 2013-05-15 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Insight into the sporulation phosphorelay: Crystal structure of the sensor domain of Bacillus subtilis histidine kinase, KinD.
Protein Sci., 22, 2013
|
|
4JGO
 
 | | The crystal structure of sporulation kinase d sensor domain from Bacillus subtilis subsp. | | Descriptor: | GLYCEROL, PYRUVIC ACID, Sporulation kinase D | | Authors: | Wu, R, Schiffer, M, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-03-01 | | Release date: | 2013-05-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Insight into the sporulation phosphorelay: Crystal structure of the sensor domain of Bacillus subtilis histidine kinase, KinD.
Protein Sci., 22, 2013
|
|
4JGR
 
 | | The crystal structure of sporulation kinase D mutant sensor domain, R131A, from Bacillus subtilis subsp at 2.4A resolution | | Descriptor: | ACETIC ACID, GLYCEROL, Sporulation kinase D | | Authors: | Wu, R, Schiffer, M, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-03-01 | | Release date: | 2013-05-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Insight into the sporulation phosphorelay: Crystal structure of the sensor domain of Bacillus subtilis histidine kinase, KinD.
Protein Sci., 22, 2013
|
|
4JGP
 
 | | The crystal structure of sporulation kinase D sensor domain from Bacillus subtilis subsp in complex with pyruvate at 2.0A resolution | | Descriptor: | PYRUVIC ACID, Sporulation kinase D | | Authors: | Wu, R, Schiffer, M, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-03-01 | | Release date: | 2013-05-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Insight into the sporulation phosphorelay: Crystal structure of the sensor domain of Bacillus subtilis histidine kinase, KinD.
Protein Sci., 22, 2013
|
|
4WKS
 
 | | n-Alkylboronic Acid Inhibitors Reveal Determinants of Ligand Specificity in the Quorum-Quenching and Siderophore Biosynthetic Enzyme PvdQ | | Descriptor: | Acyl-homoserine lactone acylase PvdQ, ethylboronic acid | | Authors: | Wu, R, Clevenger, D.K, Fast, W, Liu, D. | | Deposit date: | 2014-10-03 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.629 Å) | | Cite: | n-Alkylboronic Acid Inhibitors Reveal Determinants of Ligand Specificity in the Quorum-Quenching and Siderophore Biosynthetic Enzyme PvdQ.
Biochemistry, 53, 2014
|
|
3U27
 
 | | Crystal structure of ethanolamine utilization protein EutL from Leptotrichia buccalis C-1013-b | | Descriptor: | CALCIUM ION, GLYCEROL, Microcompartments protein, ... | | Authors: | Wu, R, Gu, M, Kerfeld, C.A, Salmeen, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-10-01 | | Release date: | 2012-02-08 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (1.852 Å) | | Cite: | Crystal structure of ethanolamine utilization protein EutL from Leptotrichia buccalis C-1013-b
To be Published
|
|
3K2N
 
 | |
5CVD
 
 | |
