6UHE
 
 | |
3C85
 
 | |
6WBT
 
 | | 2.52 Angstrom Resolution Crystal Structure of 6-phospho-alpha-glucosidase from Gut Microorganisms in Complex with NAD and Glucose-6-phosphate | | Descriptor: | 1,2-ETHANEDIOL, 6-O-phosphono-alpha-D-glucopyranose, MANGANESE (II) ION, ... | | Authors: | Wu, R, Kim, Y, Endres, M, Joachimiak, J. | | Deposit date: | 2020-03-27 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | 2.52 Angstrom Resolution Crystal Structure of 6-phospho-alpha-glucosidase from Gut Microorganisms in Complex with NAD and Glucose-6-phosphate
To Be Published
|
|
3DLP
 
 | | 4-Chlorobenzoyl-CoA Ligase/Synthetase, Mutant D402P, bound to 4CB | | Descriptor: | 4-CHLORO-BENZOIC ACID, 4-Chlorobenzoate CoA Ligase/Synthetase | | Authors: | Wu, R, Cao, J, Reger, A.S, Lu, X, Gulick, A.M, Dunaway-Mariano, D. | | Deposit date: | 2008-06-28 | | Release date: | 2009-04-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The mechanism of domain alternation in the acyl-adenylate forming ligase superfamily member 4-chlorobenzoate: coenzyme A ligase
Biochemistry, 48, 2009
|
|
3BK5
 
 | |
3H2Z
 
 | |
6VC6
 
 | | 2.1 Angstrom Resolution Crystal Structure of 6-phospho-alpha-glucosidase from Gut Microorganisms in Complex with NAD and Mn2+ | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, 6-phospho-alpha-glucosidase, GLYCEROL, ... | | Authors: | Wu, R, Kim, Y, Endres, M, Joachimiak, J. | | Deposit date: | 2019-12-20 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.133 Å) | | Cite: | 2.1 Angstrom Resolution Crystal Structure of 6-phospho-alpha-glucosidase from Gut Microorganisms in Complex with NAD and Mn2+
To Be Published
|
|
2I5U
 
 | | Crystal structure of DnaD domain protein from Enterococcus faecalis. Structural genomics target APC85179 | | Descriptor: | DnaD domain protein, MAGNESIUM ION | | Authors: | Wu, R, Zhang, R, Bargassa, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-08-25 | | Release date: | 2006-11-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.5 A crystal structure of a DnaD domain protein from Enterococcus Faecalis
To be Published, 2006
|
|
1RLJ
 
 | | Structural Genomics, a Flavoprotein NrdI from Bacillus subtilis | | Descriptor: | FLAVIN MONONUCLEOTIDE, IODIDE ION, NrdI protein | | Authors: | Wu, R, Zhang, R, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-11-25 | | Release date: | 2004-07-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 1.5A crystal structure of a thioredoxin-like protein NrdI from Bacillus subtilis
To be Published
|
|
4EW7
 
 | | The crystal structure of conjugative transfer PAS_like domain from Salmonella enterica subsp. enterica serovar Typhimurium | | Descriptor: | ACETIC ACID, CHLORIDE ION, Conjugative transfer: regulation, ... | | Authors: | Wu, R, Jedrzejczak, R.P, Brown, R.N, Cort, J.R, Heffron, F, Nakayasu, E.S, Adkins, J.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Program for the Characterization of Secreted Effector Proteins (PCSEP) | | Deposit date: | 2012-04-26 | | Release date: | 2012-09-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The crystal structure of conjugative transfer PAS_like domain from Salmonella enterica subsp. enterica serovar Typhimurium
To be Published
|
|
1RZ2
 
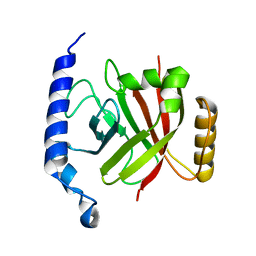 | | 1.6A crystal structure of the protein BA4783/Q81L49 (similar to sortase B) from Bacillus anthracis. | | Descriptor: | conserved hypothetical protein BA4783 | | Authors: | Wu, R, Zhang, R, Gornicki, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-12-23 | | Release date: | 2004-07-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of sortase B from Staphylococcus aureus and Bacillus anthracis reveal catalytic amino acid triad in the active site.
Structure, 12, 2004
|
|
3K32
 
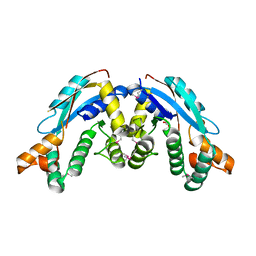 | |
4ZSW
 
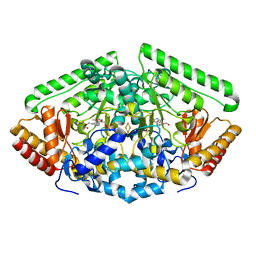 | | Pig Brain GABA-AT inactivated by (E)-(1S,3S)-3-Amino-4-fluoromethylenyl-1-cyclopentanoic acid | | Descriptor: | (1S)-4-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]cyclopent-3-ene-1,3-dicarboxylic acid, 4-aminobutyrate aminotransferase, mitochondrial, ... | | Authors: | Wu, R, Lee, H, Le, H.V, Doud, E, Sanishvili, R, Compton, P, Kelleher, N.L, Silverman, R.B, Liu, D. | | Deposit date: | 2015-05-14 | | Release date: | 2015-07-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of Inactivation of GABA Aminotransferase by (E)- and (Z)-(1S,3S)-3-Amino-4-fluoromethylenyl-1-cyclopentanoic Acid.
Acs Chem.Biol., 10, 2015
|
|
4ZSY
 
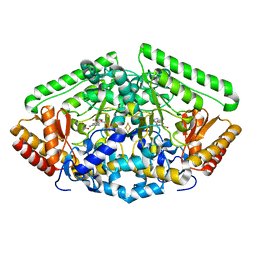 | | Pig Brain GABA-AT inactivated by (Z)-(1S,3S)-3-Amino-4-fluoromethylenyl-1-cyclopentanoic acid. | | Descriptor: | (1S)-4-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]cyclopent-3-ene-1,3-dicarboxylic acid, 4-aminobutyrate aminotransferase, mitochondrial, ... | | Authors: | Wu, R, Lee, H, Le, H.V, Doud, E, Sanishvili, R, Compton, P, Kelleher, N.L, Silverman, R.B, Liu, D. | | Deposit date: | 2015-05-14 | | Release date: | 2015-07-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of Inactivation of GABA Aminotransferase by (E)- and (Z)-(1S,3S)-3-Amino-4-fluoromethylenyl-1-cyclopentanoic Acid.
Acs Chem.Biol., 10, 2015
|
|
4Y0I
 
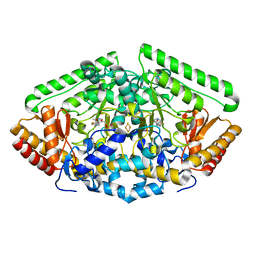 | | GABA-aminotransferase inactivated by conformationally-restricted inactivator | | Descriptor: | 4-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]THIOPHENE-2-CARBOXYLIC ACID, 4-aminobutyrate aminotransferase, mitochondrial, ... | | Authors: | Wu, R, Sanishvili, R, Lee, H.V, Dustin, D.H, Emma, D, Neil, K, Silverman, R.B, Liu, D. | | Deposit date: | 2015-02-06 | | Release date: | 2016-02-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.662 Å) | | Cite: | GABA-aminotransferase inactivated by conformationally-restricted inactivator
To Be Published
|
|
4ZGJ
 
 | | Double Mutant H80A/H81A of Fe-Type Nitrile Hydratase from Comamonas testosteroni Ni1 | | Descriptor: | FE (III) ION, Nitrile hydratase alpha subunit, Nitrile hydratase beta subunit | | Authors: | Wu, R, Martinez, S, Holz, R, Liu, D. | | Deposit date: | 2015-04-23 | | Release date: | 2015-07-01 | | Last modified: | 2015-07-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analyzing the catalytic role of active site residues in the Fe-type nitrile hydratase from Comamonas testosteroni Ni1.
J.Biol.Inorg.Chem., 20, 2015
|
|
2P7J
 
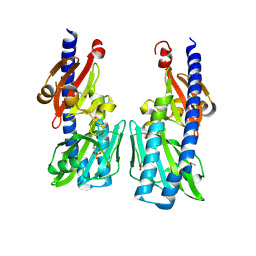 | |
4ZGD
 
 | | Mutant R157A of Fe-Type Nitrile Hydratase from Comamonas testosteroni Ni1 | | Descriptor: | FE (III) ION, Nitrile hydratase alpha subunit, Nitrile hydratase beta subunit | | Authors: | Wu, R, Martinez, S, Holz, R, Liu, D. | | Deposit date: | 2015-04-22 | | Release date: | 2015-07-01 | | Last modified: | 2015-07-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Analyzing the catalytic role of active site residues in the Fe-type nitrile hydratase from Comamonas testosteroni Ni1.
J.Biol.Inorg.Chem., 20, 2015
|
|
2PPW
 
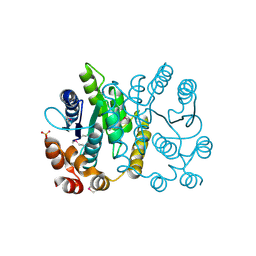 | |
4ZGE
 
 | | Double Mutant H80W/H81W of Fe-Type Nitrile Hydratase from Comamonas testosteroni Ni1 | | Descriptor: | FE (III) ION, Nitrile hydratase alpha subunit, Nitrile hydratase beta subunit | | Authors: | Wu, R, Martinez, S, Holz, R, Liu, D. | | Deposit date: | 2015-04-22 | | Release date: | 2015-07-01 | | Last modified: | 2015-07-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Analyzing the catalytic role of active site residues in the Fe-type nitrile hydratase from Comamonas testosteroni Ni1.
J.Biol.Inorg.Chem., 20, 2015
|
|
2OQW
 
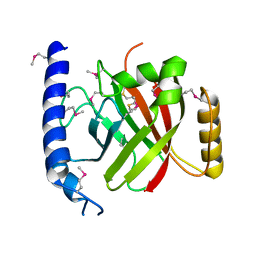 | | The crystal structure of sortase B from B.anthracis in complex with AAEK1 | | Descriptor: | Sortase B | | Authors: | Wu, R, Zhang, R, Marresso, A.W, Schneewind, O, Joachimiak, A. | | Deposit date: | 2007-02-01 | | Release date: | 2007-06-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Activation of inhibitors by sortase triggers irreversible modification of the active site.
J.Biol.Chem., 282, 2007
|
|
2OQZ
 
 | | The crystal structure of sortase B from B.anthracis in complex with AAEK2 | | Descriptor: | ACETIC ACID, Sortase B | | Authors: | Wu, R, Zhang, R, Maresso, A.W, Schneewind, O, Joachimiak, A. | | Deposit date: | 2007-02-01 | | Release date: | 2007-06-19 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Activation of inhibitors by sortase triggers irreversible modification of the active site.
J.Biol.Chem., 282, 2007
|
|
2OT9
 
 | |
2QSX
 
 | | Crystal structure of putative transcriptional regulator LysR From Vibrio parahaemolyticus | | Descriptor: | Putative transcriptional regulator, LysR family, SULFATE ION | | Authors: | Wu, R, Abdullah, J, Binkowski, T.A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-31 | | Release date: | 2007-09-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | The Crystal Structure of Putative Transcriptional Regulator LysR From Vibrio parahaemolyticus.
To be Published
|
|
2QW0
 
 | | 4-Chlorobenzoyl-CoA Ligase/Synthetase, I303A mutation, bound to 3,4 Dichlorobenzoate | | Descriptor: | 3,4-dichlorobenzoate, 4-Chlorobenzoate CoA Ligase | | Authors: | Wu, R, Reger, A.S, Cao, J, Gulick, A.M, Dunaway-Mariano, D. | | Deposit date: | 2007-08-09 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Rational redesign of the 4-chlorobenzoate binding site of 4-chlorobenzoate: coenzyme a ligase for expanded substrate range.
Biochemistry, 46, 2007
|
|
