6DBJ
 
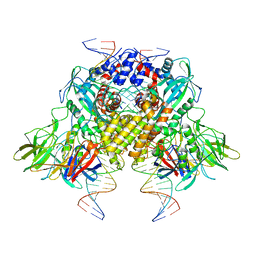 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS nicked DNA intermediates | | Descriptor: | CALCIUM ION, Forward stand of RSS signal end, Forward strand of coding flank, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
4ZGY
 
 | |
1NX7
 
 | |
1DS9
 
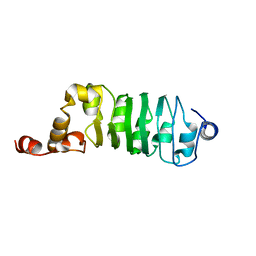 | | SOLUTION STRUCTURE OF CHLAMYDOMONAS OUTER ARM DYNEIN LIGHT CHAIN 1 | | Descriptor: | OUTER ARM DYNEIN | | Authors: | Wu, H.W, Maciejewski, M.W, Marintchev, A, Benashski, S.E, Mullen, G.P, King, S.M. | | Deposit date: | 2000-01-07 | | Release date: | 2000-07-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a dynein motor domain associated light chain.
Nat.Struct.Biol., 7, 2000
|
|
1J0Q
 
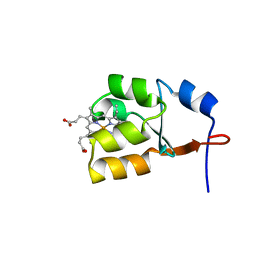 | | Solution Structure of Oxidized Bovine Microsomal Cytochrome b5 mutant V61H | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, cytochrome b5 | | Authors: | Wu, H, Huang, Z, Cao, C, Zhang, Q, Wang, Y.-H, Ma, J.-B, Xue, L.-L. | | Deposit date: | 2002-11-20 | | Release date: | 2003-08-12 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the oxidized bovine microsomal cytochrome b5 mutant V61H
Biochem.Biophys.Res.Commun., 307, 2003
|
|
3GD9
 
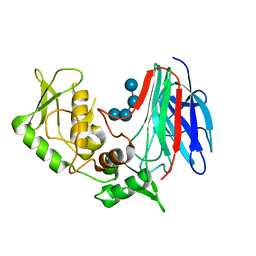 | | Crystal structure of laminaripentaose-producing beta-1,3-glucanase in complex with laminaritetraose | | Descriptor: | Laminaripentaose-producing beta-1,3-guluase (LPHase), beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-alpha-D-glucopyranose | | Authors: | Wu, H.M, Hsu, M.T, Liu, S.W, Lai, C.C, Li, Y.K, Wang, W.C. | | Deposit date: | 2009-02-23 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure, mechanistic action, and essential residues of a GH-64 enzyme, laminaripentaose-producing beta-1,3-glucanase.
J.Biol.Chem., 284, 2009
|
|
3GD0
 
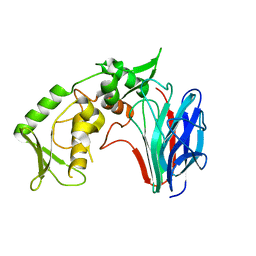 | | Crystal structure of laminaripentaose-producing beta-1,3-glucanase | | Descriptor: | Laminaripentaose-producing beta-1,3-guluase (LPHase) | | Authors: | Wu, H.M, Hsu, M.T, Liu, S.W, Lai, C.C, Li, Y.K, Wang, W.C. | | Deposit date: | 2009-02-23 | | Release date: | 2009-07-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure, mechanistic action, and essential residues of a GH-64 enzyme, laminaripentaose-producing beta-1,3-glucanase.
J.Biol.Chem., 284, 2009
|
|
2FJ4
 
 | |
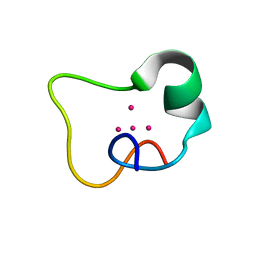
2FJ5
 
 | |
1WIQ
 
 | |
4DJH
 
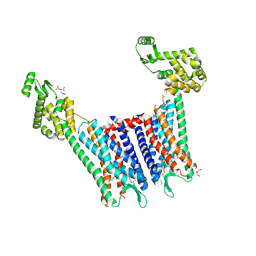 | | Structure of the human kappa opioid receptor in complex with JDTic | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (3R)-7-hydroxy-N-{(2S)-1-[(3R,4R)-4-(3-hydroxyphenyl)-3,4-dimethylpiperidin-1-yl]-3-methylbutan-2-yl}-1,2,3,4-tetrahydroisoquinoline-3-carboxamide, CITRIC ACID, ... | | Authors: | Wu, H, Wacker, D, Katritch, V, Mileni, M, Han, G.W, Vardy, E, Liu, W, Thompson, A.A, Huang, X.P, Carroll, F.I, Mascarella, S.W, Westkaemper, R.B, Mosier, P.D, Roth, B.L, Cherezov, V, Stevens, R.C, GPCR Network (GPCR) | | Deposit date: | 2012-02-01 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | tructure of the human kappa-opioid receptor in complex with JDTic
Nature, 485, 2012
|
|
2JOK
 
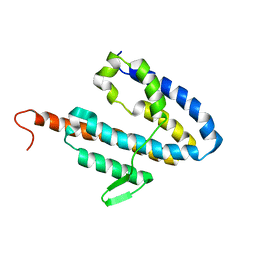 | | NMR structure of the catalytic domain of guanine nucleotide exchange factor BopE from Burkholderia pseudomallei | | Descriptor: | Putative G-nucleotide exchange factor | | Authors: | Wu, H, Upadhyay, A, Williams, C, Galyov, E.E, van den Elsen, J.M.H, Bagby, S. | | Deposit date: | 2007-03-14 | | Release date: | 2007-09-18 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | The guanine-nucleotide-exchange factor BopE from Burkholderia pseudomallei adopts a compact version of the Salmonella SopE/SopE2 fold and undergoes a closed-to-open conformational change upon interaction with Cdc42
Biochem.J., 411, 2008
|
|
2JOL
 
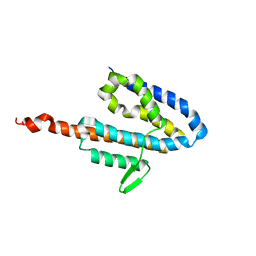 | | Average NMR structure of the catalytic domain of guanine nucleotide exchange factor BopE from Burkholderia pseudomallei | | Descriptor: | Putative G-nucleotide exchange factor | | Authors: | Wu, H, Upadhyay, A, Williams, C, Galyov, E.E, van den Elsen, J.M.H, Bagby, S. | | Deposit date: | 2007-03-14 | | Release date: | 2007-03-27 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | The guanine-nucleotide-exchange factor BopE from Burkholderia pseudomallei adopts a compact version of the Salmonella SopE/SopE2 fold and undergoes a closed-to-open conformational change upon interaction with Cdc42
Biochem.J., 411, 2008
|
|
6DBW
 
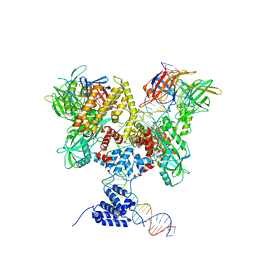 | | Cryo-EM structure of RAG in complex with 12-RSS substrate DNA | | Descriptor: | CALCIUM ION, Forward strand of 12-RSS substrate DNA, Recombination activating gene 1 - MBP chimera, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DBX
 
 | | Cryo-EM structure of RAG in complex with 12-RSS substrate DNA | | Descriptor: | CALCIUM ION, Forward strand of 12-RSS substrate DNA, Recombination activating gene 1 - MBP chimera, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
1XDX
 
 | |
1SH4
 
 | |
7E5Y
 
 | | Molecular basis for neutralizing antibody 2B11 targeting SARS-CoV-2 RBD | | Descriptor: | 2B11 Fab Heavy chain, 2B11 Fab Light chain, Spike protein S1 | | Authors: | Wu, H, Yu, F, Wang, Q.S, Zhou, H, Wang, W.W, Zhao, T, Pan, Y.B, Yang, X.M. | | Deposit date: | 2021-02-21 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Screening of potent neutralizing antibodies against SARS-CoV-2 using convalescent patients-derived phage-display libraries.
Cell Discov, 7, 2021
|
|
5WOD
 
 | |
5UNA
 
 | | Fragment of 7SK snRNA methylphosphate capping enzyme | | Descriptor: | 7SK snRNA methylphosphate capping enzyme, S-ADENOSYL-L-HOMOCYSTEINE, unidentified peptide section/fragment | | Authors: | Wu, H, Tempel, W, Dombrovski, L, McCarthy, A.A, Loppnau, P, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-01-30 | | Release date: | 2017-03-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Fragment of 7SK snRNA methylphosphate capping enzyme
To Be Published
|
|
2P89
 
 | |
2P0W
 
 | | Human histone acetyltransferase 1 (HAT1) | | Descriptor: | ACETAMIDE, ACETATE ION, ACETYL COENZYME *A, ... | | Authors: | Wu, H, Min, J, Zeng, H, Loppnau, P, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-03-01 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of human histone acetyltransferase 1 (HAT1) in complex with acetylcoenzyme A and histone peptide H4
To be Published
|
|
2O71
 
 | | Crystal structure of RAIDD DD | | Descriptor: | Death domain-containing protein CRADD | | Authors: | Wu, H, Park, H. | | Deposit date: | 2006-12-09 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of RAIDD Death Domain Implicates Potential Mechanism of PIDDosome Assembly
J.Mol.Biol., 357, 2006
|
|
1K4A
 
 | | STRUCTURE OF AGAA RNA TETRALOOP, NMR, 20 STRUCTURES | | Descriptor: | 5'-R(*GP*GP*UP*UP*CP*AP*GP*AP*AP*GP*AP*AP*CP*C)-3' | | Authors: | Wu, H, Yang, P.K, Butcher, S.E, Kang, S, Chanfreau, G, Feigon, J. | | Deposit date: | 2001-10-07 | | Release date: | 2001-12-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A novel family of RNA tetraloop structure forms the recognition site for Saccharomyces cerevisiae RNase III.
EMBO J., 20, 2001
|
|
1K4B
 
 | | STRUCTURE OF AGUU RNA TETRALOOP, NMR, 20 STRUCTURES | | Descriptor: | 5'-R(*GP*GP*UP*UP*CP*AP*GP*UP*UP*GP*AP*AP*CP*C)-3' | | Authors: | Wu, H, Yang, P.K, Butcher, S.E, Kang, S, Chanfreau, G, Feigon, J. | | Deposit date: | 2001-10-07 | | Release date: | 2001-12-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A novel family of RNA tetraloop structure forms the recognition site for Saccharomyces cerevisiae RNase III.
EMBO J., 20, 2001
|
|
