7JN5
 
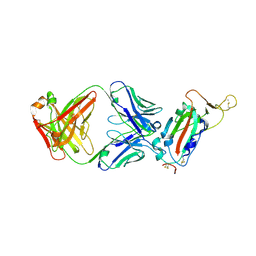 | | Crystal structure of SARS-CoV receptor binding domain in complex with human antibody CR3022 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CR3022 heavy chain, ... | | Authors: | Wu, N.C, Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2020-08-03 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.705 Å) | | Cite: | A natural mutation between SARS-CoV-2 and SARS-CoV determines neutralization by a cross-reactive antibody.
Plos Pathog., 16, 2020
|
|
2JP9
 
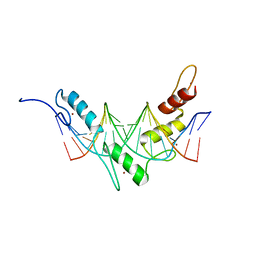 | | Structure of the Wilms Tumor Suppressor Protein Zinc Finger Domain Bound to DNA | | Descriptor: | DNA (5'-D(P*DCP*DGP*DCP*DGP*DGP*DGP*DGP*DGP*DCP*DGP*DTP*DCP*DTP*DGP*DCP*DGP*DC)-3'), DNA (5'-D(P*DGP*DCP*DGP*DCP*DAP*DGP*DAP*DCP*DGP*DCP*DCP*DCP*DCP*DCP*DGP*DCP*DG)-3'), Wilms tumor 1, ... | | Authors: | Stoll, R, Lee, B.M, Debler, E.W, Laity, J.H, Wilson, I.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2007-04-30 | | Release date: | 2007-10-30 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structure of the wilms tumor suppressor protein zinc finger domain bound to DNA
J.Mol.Biol., 372, 2007
|
|
2I27
 
 | |
7JMP
 
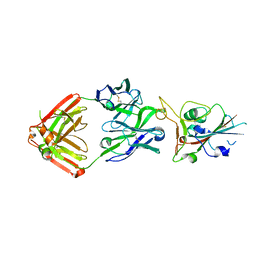 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody COVA2-39 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COVA2-39 heavy chain, COVA2-39 light chain, ... | | Authors: | Wu, N.C, Yuan, M, Liu, H, Zhu, X, Wilson, I.A. | | Deposit date: | 2020-08-02 | | Release date: | 2020-08-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.712 Å) | | Cite: | An Alternative Binding Mode of IGHV3-53 Antibodies to the SARS-CoV-2 Receptor Binding Domain.
Cell Rep, 33, 2020
|
|
7K75
 
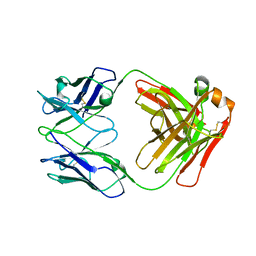 | |
7K76
 
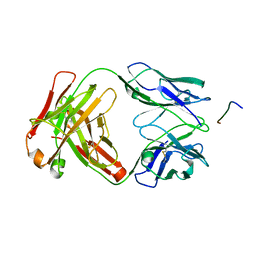 | |
7LOP
 
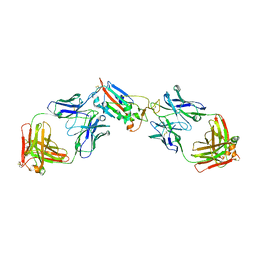 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with antibodies CV05-163 and CR3022 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CR3022 Fab heavy chain, CR3022 Fab light chain, ... | | Authors: | Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2021-02-10 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.246 Å) | | Cite: | Structural and functional ramifications of antigenic drift in recent SARS-CoV-2 variants.
Science, 373, 2021
|
|
7KN7
 
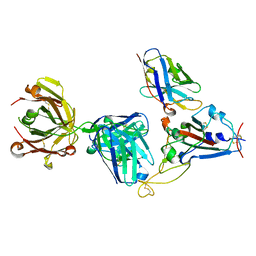 | | Crystal structure of SARS-CoV-2 receptor binding domain complexed with nanobody VHH W and antibody Fab CC12.3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CC12.3 Fab heavy chain, CC12.3 Fab light chain, ... | | Authors: | Liu, H, Yuan, M, Zhu, X, Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-11-04 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape.
Science, 371, 2021
|
|
7LA7
 
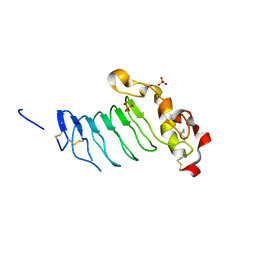 | | O6 variable lymphocyte receptor ectodomain | | Descriptor: | PHOSPHATE ION, variable lymphocyte receptor O6 | | Authors: | Bernard, S.M, Wilson, I.A. | | Deposit date: | 2021-01-06 | | Release date: | 2021-07-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.55100727 Å) | | Cite: | Novel lamprey antibody recognizes terminal sulfated galactose epitopes on mammalian glycoproteins.
Commun Biol, 4, 2021
|
|
7LA8
 
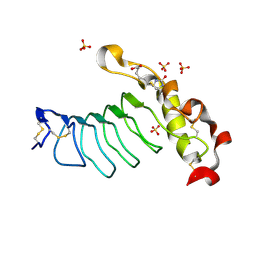 | |
2GK0
 
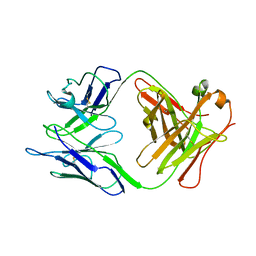 | |
7LMO
 
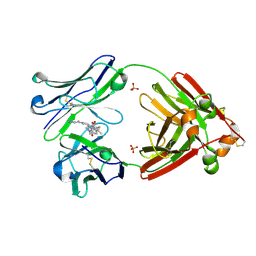 | | Structure of full-length human lambda-6A light chain JTO in complex with stabilizer 34 [3-(2-(7-(diethylamino)-4-methyl-2-oxo-2H-chromen-3-yl)ethyl)-7-(1H-imidazole-5-carbonyl)-1,3,7-triazaspiro[4.4]nonane-2,4-dione] | | Descriptor: | (5~{R})-3-[2-[7-(diethylamino)-4-methyl-2-oxidanylidene-chromen-3-yl]ethyl]-7-(1~{H}-imidazol-4-ylcarbonyl)-1,3,7-triazaspiro[4.4]nonane-2,4-dione, JTO light chain, PHOSPHATE ION | | Authors: | Yan, N.L, Wilson, I.A, Kelly, J.W. | | Deposit date: | 2021-02-05 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Discovery of Potent Coumarin-Based Kinetic Stabilizers of Amyloidogenic Immunoglobulin Light Chains Using Structure-Based Design.
J.Med.Chem., 64, 2021
|
|
7LMN
 
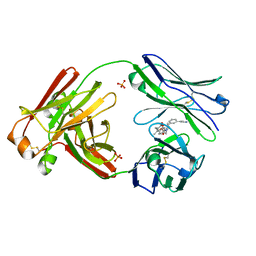 | |
7LMR
 
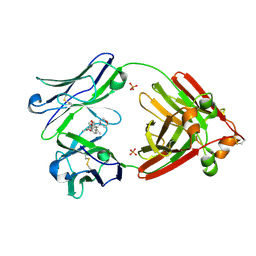 | |
7LMP
 
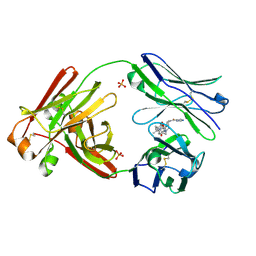 | | Structure of full-length human lambda-6A light chain JTO in complex with stabilizer 36 [3-(2-(7-(diethylamino)-4-methyl-2-oxo-2H-chromen-3-yl)ethyl)-8-(1H-imidazole-4-carbonyl)-1,3,8-triazaspiro[4.5]decane-2,4-dione] | | Descriptor: | 3-[2-[7-(diethylamino)-4-methyl-2-oxidanylidene-chromen-3-yl]ethyl]-8-(1~{H}-imidazol-5-ylcarbonyl)-1,3,8-triazaspiro[4.5]decane-2,4-dione, JTO light chain, PHOSPHATE ION | | Authors: | Yan, N.L, Wilson, I.A, Kelly, J.W. | | Deposit date: | 2021-02-05 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Discovery of Potent Coumarin-Based Kinetic Stabilizers of Amyloidogenic Immunoglobulin Light Chains Using Structure-Based Design.
J.Med.Chem., 64, 2021
|
|
7LMQ
 
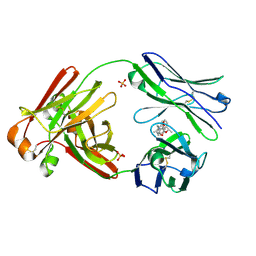 | |
7LQ7
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with antibodies CV503 and COVA1-16 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COVA1-16 heavy chain, COVA1-16 light chain, ... | | Authors: | Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2021-02-13 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Bispecific antibodies targeting distinct regions of the spike protein potently neutralize SARS-CoV-2 variants of concern.
Sci Transl Med, 13, 2021
|
|
2I24
 
 | |
2I26
 
 | |
2GJZ
 
 | |
2KA0
 
 | | NMR structure of the protein TM1367 | | Descriptor: | uncharacterized protein TM1367 | | Authors: | Mohanty, B, Pedrini, B, Serrano, P, Geralt, M, Horst, R, Herrmann, T, Wilson, I.A, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-10-27 | | Release date: | 2009-01-13 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Comparison of NMR and crystal structures for the proteins TM1112 and TM1367.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2JPA
 
 | | Structure of the Wilms Tumor Suppressor Protein Zinc Finger Domain Bound to DNA | | Descriptor: | DNA (5'-D(P*DCP*DAP*DGP*DAP*DCP*DGP*DCP*DCP*DCP*DCP*DCP*DGP*DCP*DG)-3'), DNA (5'-D(P*DCP*DGP*DCP*DGP*DGP*DGP*DGP*DGP*DCP*DGP*DTP*DCP*DTP*DG)-3'), Wilms tumor 1, ... | | Authors: | Stoll, R, Lee, B.M, Debler, E.W, Laity, J.H, Wilson, I.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2007-05-01 | | Release date: | 2007-10-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the wilms tumor suppressor protein zinc finger domain bound to DNA
J.Mol.Biol., 372, 2007
|
|
2KL4
 
 | | NMR structure of the protein NB7804A | | Descriptor: | BH2032 protein | | Authors: | Mohanty, B, Geralt, M, Augustyniak, W, Serrano, P, Horst, R, Wilson, I.A, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2009-06-30 | | Release date: | 2009-07-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the protein NB7804A from Bacillus Halodurans
To be Published
|
|
2KYZ
 
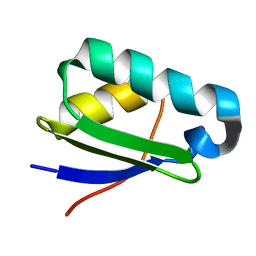 | | NMR structure of heavy metal binding protein TM0320 from Thermotoga maritima | | Descriptor: | Heavy metal binding protein | | Authors: | Jaudzems, K, Wahab, A, Serrano, P, Geralt, M, Wuthrich, K, Wilson, I.A, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2010-06-09 | | Release date: | 2010-07-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure of heavy metal binding protein TM0320 from Thermotoga maritima
To be Published
|
|
2KZC
 
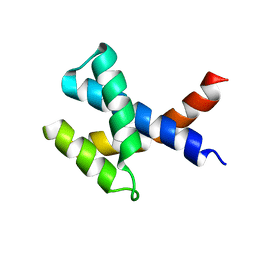 | | Solution NMR structure of the protein YP_510488.1 | | Descriptor: | Uncharacterized protein | | Authors: | Mohanty, B, Serrano, P, Geralt, M, Horst, R, Wilson, I.A, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2010-06-15 | | Release date: | 2010-07-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the protein YP_510488.1
To be Published
|
|
